Abstract
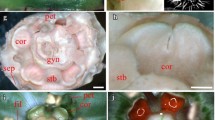
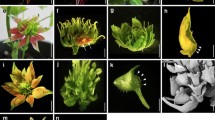
In higher eudicotyledonous angiosperms the floral organs are typically arranged in four different whorls, containing sepals, petals, stamens and carpels. According to the ABC model, the identity of these organs is specified by floral homeotic genes of class A, A+B, B+C and C, respectively. In contrast to the sepal and petal whorls of eudicots, the perianths of many plants from the Liliaceae family have two outer whorls of almost identical petaloid organs, called tepals. To explain the Liliaceae flower morphology, van Tunen et al. (1993) proposed a modified ABC model, exemplified with tulip. According to this model, class B genes are not only expressed in whorls 2 and 3, but also in whorl 1. Thus the organs of both whorls 1 and 2 express class A plus class B genes and, therefore, get the same petaloid identity. To test this modified ABC model we have cloned and characterized putative class B genes from tulip. Two DEF- and one GLO-like gene were identified, named TGDEFA, TGDEFB and TGGLO. Northern hybridization analysis showed that all of these genes are expressed in whorls 1, 2 and 3 (outer and inner tepals and stamens), thus corroborating the modified ABC model. In addition, these experiments demonstrated that TGGLO is also weakly expressed in carpels, leaves, stems and bracts. Gel retardation assays revealed that TGGLO alone binds to DNA as a homodimer. In contrast, TGDEFA and TGDEFB cannot homodimerize, but make heterodimers with PI. Homodimerization of GLO-like protein has also been reported for lily, suggesting that this phenomenon is conserved within Liliaceae plants or even monocot species.
Similar content being viewed by others
References
Albert, V.A., Gustaffson, M.H.G. and Di Laurenzio, L. 1998. Ontogenetic systematics, molecular developmental genetics, and the angiosperm petal. In: D.E. Soltis, P.S. Soltis and J.J. Doyle (Eds.) Molecular Systematics of Plants II, Kluwer Academic Publishers, Boston, MA/Dordrecht, Netherlands, pp. 349-374.
Ambrose, B.A., Lerner, D.R., Ciceri, P., Padilla, C.M., Yanofsky, M.F. and Schmidt, R.J. 2000. Molecular and genetic analyses of the Silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Mol. Cell 5: 569-579.
Baum, D.A. 1998. The evolution of plant development. Curr. Opin. Plant Biol. 1: 79-86.
Baum, D.A. and Whitlock, B.A. 1999. Genetic clues to petal evolution. Curr. Biol. 9: R525-R527.
Chung, Y.Y., Kim, S.R., Kang, H.G., Noh, Y.S., Park, M.C., Finkel, D. and An, G. 1995. Characterization of two rice MADS box genes homologous to GLOBOSA. Plant Sci. 109: 45-56.
Coen, E.S. and Meyerowitz, E.M. 1991. The war of the whorls: genetic interactions controlling flower development. Nature 353: 31-37.
Dahlgren, R.M.T., Clifford, H.T. and Yeo, P.F. 1985. In: Dahlgren, R.M.T., Clifford, H.T., Yeo, P.F. (Eds.) The Families of the Monocotyledons, Springer-Verlag, Berlin/Heidelberg, pp. 65.
Egea-Cortines, M., Saedler, H. and Sommer, H. 1999. Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J. 18: 5370-5379.
Frohman, M.A., Dush, M.K. and Martin, G.R. 1988. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc. Natl. Acad. Sci. USA 85: 8998-9002.
Goto, K. and Meyerowitz, E.M. 1994. Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 8: 1548-1560.
Honma, T. and Goto, K. 2001. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409: 525-529.
Jack, T., Brockman, L.L. and Meyerowitz, E.M. 1992. The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68: 683-697.
Kang, H.G., Jeon, J.S., Lee, S. and An, G. 1998. Identification of class B and class C floral organ identity genes from rice plants. Plant Mol. Biol. 38: 1021-1029.
Kisaka, H., Kisaka, M. and Kameya, T. 1996. Characterization of interfamilial somatic hybrids between 5-methyltryptophanresistant (5MT-resistant) rice (Oryza sativa L.) and 5MTsensitive carrot (Daucus carota L.): expression of resistance to 5MT by somatic hybrids. Breed. Sci. 46: 221-226.
Kramer, E.M. and Irish, V.F. 2000. Evolution of the petal and stamen developmental programs: evidence from comparative studies of the lower eudicots and basal angiosperms. Int. J. Plant Sci. 161 (Suppl.): S29-S40.
Kramer, E.M., Dorit, R.L. and Irish, V.F. 1998. Molecular evolution of genes controlling petal and stamen development: duplication and divergence within the APETALA3 and PISTILLATA MADSbox gene lineages. Genetics 149: 765-783.
Krizek, B.A. and Meyerowitz, E.M. 1996. The Arabidopsis homeotic genes APETALA3 and PISTILLATA are sufficient to provide the B class organ identity function. Development 122: 11-22.
Kyozuka, J., Kobayashi, T., Morita, M. and Shimamoto, K. 2000.Spatially and temporally regulated expression of rice MADS box genes with similarity to Arabidopsis class A, B and C genes. Plant Cell Physiol. 41: 710-718.
Lee, S., Jeon, J.S., Moon, Y.H. and An, G. 2000. Alteration of rice floral organ identity by ectopic expression of rice MADSbox gene. In: Proceedings of the 4th International Rice Genetics Symposium.
Moon, Y.H., Jung, J.Y., Kang, H.G. and An, G. 1999. Identification of a rice APETALA3 homologue by yeast two-hybrid screening. Plant Mol. Biol. 40: 167-177.
Münster, T., Pahnke, J., Di Rosa, A., Kim, J.T., Martin, W., Saedler, H. and Theissen, G. 1997. Floral homeotic genes were recruited from homologous MADS-box genes preexisting in the common ancestor of ferns and seed plants. Proc. Natl. Acad. Sci. USA 94: 2415-2420.
Münster, T., Wingen, L.U., Faigl, W., Werth, S., Saedler, H. and Theissen, G. 2001. Characterization of three GLOBOSA-like MADS-box genes from maize: evidence for ancient paralogy in one class of floral homeotic B-function genes of grasses. Gene 262: 1-13.
Murray, M.G. and Thompson, W.F. 1980. Rapid isolation of high molecular weight plant DNA. Nucl. Acid. Res. 8: 4321-4325.
Park, J.H., Ishikawa, Y., Yoshida, R., Kanno, A. and Kameya, T. 2003. Expression of AODEF, a B-functional MADS-box gene, in stamens and inner tepals of the dioecious species Asparagus officinalis L. Plant Mol. Biol. 51: 867-875.
Pelaz, S., Tapia-Lopez, R., Alvarez-Buylla, E.R. and Yanofsky, M.F. 2001. Conversion of leaves into petals in Arabidopsis. Curr. Biol. 11: 182-184.
Perrière, G. and Gouy, M. 1996. WWW-Query: an on-line retrieval system for biological sequence banks. Biochemie 78: 364-369.
Riechmann, J.L., Krizek, B.A. and Meyerowitz, E.M. 1996. Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA and AGAMOUS. Proc. Natl. Acad. Sci. USA 93: 4793-4798.
Sambrook, J. and Russell, D.W. 2001. Molecular Cloning: A Laboratory Manual, 3rd ed. Cold Spring Harbor Laboratory Press, Plainview, NY.
Schwarz-Sommer, Z., Hue, I., Huijser, P., Flor, P.J., Hansen, R., Tetens, F., Lönnig, W.E., Saedler, H. and Sommer, H. 1992. Characterization of the Antirrhinum floral homeotic MADS-box gene deficiens: evidence for DNA binding and autoregulation of its persistent expression throughout flower development. EMBO J. 11: 251-263.
Soltis, P.S., Soltis, D.E. and Chase, M.W. 1999. Angiosperm phylogeny inferred from multiple genes as a tool for comparative biology. Nature 402: 402-403.
Theissen, G. 2001. Development of floral organ identity: stories from the MADS house. Curr. Opin. Plant Biol. 4: 75-85.
Theissen, G. and Saedler, H. 2001. Floral quartets. Nature 409: 469-471.
Theissen, G., Kim, J. and Saedler, H. 1996. Classification and phylogeny of the MADS-box multigene family suggest defined roles of MADS-box gene subfamilies in the morphological evolution of eukaryotes. J. Mol. Evol. 43: 484-516.
Theissen, G., Becker, A., Di Rosa, A., Kanno, A., Kim, J.T., Münster, T., Winter, K.U. and Saedler, H. 2000. A short history of MADS-box genes in plants. Plant Mol. Biol. 42: 115-149.
Thompson, J.D., Higgins, D.G. and Gibson, T.J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucl. Acids. Res. 22: 4673-4680.
Tröbner, W., Ramirez, L., Motte, P., Hue, I., Huijser, P., Lönnig, W.E., Saedler, H., Sommer, H. and Schwarz-Sommer, Z. 1992. GLOBOSA: a homeotic gene which interacts with DEFICIENS in the control of Antirrhinum floral organogenesis. EMBO J. 11: 4693-4704.
Tzeng, T.Y. and Yang, C.H. 2001. A MADS box gene from lily (Lilium longiflorum) is sufficient to generate dominant negative mutation by interacting with PISTILLATA (PI) in Arabidopsis thaliana. Plant Cell Physiol. 42: 1156-1168.
van Tunen, A.J., Eikelboom, W. and Angenent, G.C. 1993. Floral organogenesis in Tulipa. Flow. Newsl. 16: 33-38.
Weigel, D. and Meyerowitz, E.M. 1994. The ABCs of floral homeotic genes. Cell 78: 203-209.
Winter, K.U., Weiser, C., Kaufmann, K., Bohne, A., Kirchner, C., Kanno, A., Saedler, H. and Theissen, G. 2002. Evolution of class B floral homeotic proteins: obligate heterodimerization originated from homodimerization. Mol. Biol. Evol. 19: 587-596.
Zachgo, S., Silva, E.A., Motte, P., Tröbner, W., Saedler, H. and Schwarz-Sommer, Z. 1995. Functional analysis of the Antirrhinum floral homeotic DEFICIENS gene in vivo and in vitro by using a temperature-sensitive mutant. Development 121: 2861-2875.
Author information
Authors and Affiliations
Additional information
these authors contributed equally to this work
Rights and permissions
About this article
Cite this article
Kanno, A., Saeki, H., Kameya, T. et al. Heterotopic expression of class B floral homeotic genes supports a modified ABC model for tulip (Tulipa gesneriana). Plant Mol Biol 52, 831–841 (2003). https://doi.org/10.1023/A:1025070827979
Issue Date:
DOI: https://doi.org/10.1023/A:1025070827979