Abstract
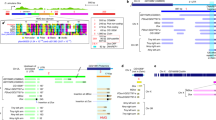
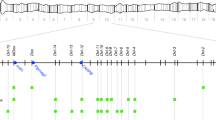
In D. melanogaster males carrying Segregation Distorter (SD) second chromosomes, sperm receiving sensitive alleles of the Responder (Rsp) locus are subject to high rates of dysfunction. The Rsp region is located in 2R immediately adjacent to the centromere in heterochromatic band 39, and covers roughly 600 kb of material, of which approximately 85 kb is comprised of several hundred copies of a 240-bp satellite DNA sequence. Cytological observations as well as molecular analysis of rearrangements which bisect h39 indicate that sensitivity of the Rsp target to SD action is also subdivisible, and sensitivities of the component pieces appear to be correlated with copy number of the 240 bp repeat. In an attempt to examine possible higher order sequence structure for these blocks, PCR using single primers derived from a canonical repeat was used to identify potential reversals of direction of tandem arrays; that is, head-to-head or tail-to-tail junctions. Surprisingly, for two different Rsp alleles, only a single such reversal product for each was identified, differing in size and sequence between alleles. Sequencing of PCR products identified diverged copies of the canonical repeats that would not have been found using the levels of DNA stringency employed in earlier studies. Examination of Southern digests and slot-blots for DNA quantification indicates that adding the estimated numbers of such diverged copies to the canonical repeat copies discovered earlier is potentially sufficient to account for the entire 600 kb Rsp region. This adds strength to the hypothesis that this extended family of repeats is in fact the target of SD-mediated sperm dysfunction. Implications of these results for understanding the evolution of repetitive DNA are also discussed.
Similar content being viewed by others
References
Brittnacher, J.G. & B. Ganetzky, 1983. On the components of Segregation distortion in Drosophila melanogaster. II. Deletion mapping and dosage analysis of the SD locus. Genetics 103: 659–673.
Brittnacher, J.G. & B. Ganetzky, 1989. On the components of Segregation Distortion in Drosophila melanogaster, IV. Construction and analysis of free duplications for the Responder locus. Genetics 121: 739–750.
Brizuela, B.J., L. Elfring et al., 1994. Genetic analysis of the brahma gene of Drosophila melanogaster and polytene chromosome subdivisions 72AB. Genetics 137: 803–813.
Cabot, E.L., P. Doshi et al., 1993. Population genetics of tandem repeats in centromeric heterochromatin: unequal crossing over and chromosomal divergence at the Responder locus of Drosophila melanogaster. Genetics 135: 477–487.
Church, G.M. & W. Gilbert, 1984. Genomic sequencing. Proc. Natl. Acad. Sci. USA 81: 1991-1995.
Coelho, P.A., D. Nurminsky et al., 1996. Identification of Porto-1, a new repeated sequence that localises close to the centromere of chromosome 2 of Drosophila melanogaster. Chromosoma 105: 211–222.
Cornish, E.C., S.A. Beckham et al., 1998. Southern hybridization revisited; probe/target DNA interaction is affected by the choice of hybridization buffer. Biotechniques 25: 948–950, 952, 954.
Doshi, P., S. Kaushal, C. Benyajati & C.-I. Wu, 1991. Molecular analysis of the responder satellite DNA in Drosophila melanogaster: DNA bending, nucleosome structure, and Rsp-binding proteins. Mol. Biol. Evol. 8: 721–741.
Gatti, M. & S. Pimpinelli, 1992. Functional elements in Drosophila melanogaster heterochromatin. Annu. Rev. Genet. 26: 239–275.
Kuh, J., K. Houtchens et al., 1995. Pulsed-field gel analysis of the 2R heterochromatic locus, Responder. A. Conf. Dros. Res. 36: 200A.
Le, M.H., D. Duricka et al., 1995. Islands of complex DNA are widespread in Drosophila centric heterochromatin. Genetics 141: 283–303.
Lindsley, D. & G. Zimm, 1992. The Genome of Drosophila melanogaster. Academic Press, San Diego.
Lyttle, T.W., 1991. Segregation distorters. Annu. Rev. Genet 25: 511–557.
Lyttle, T.W., 1993. Cheaters sometimes prosper: distortion of Mendelian segregation by meiotic drive. Trends Genet. 9: 205–210.
Merrill, C. & B. Ganetzky, 1999. Immunolocalization of Sdencoded mutant RanGAP in spermatogenesis. A. Dros. Res. Conf. 40.
Merrill, C., L. Bayraktaroglu et al., 1999. Truncated RanGAP encoded by the Segregation Distorter locus of Drosophila [see comments]. Science 283: 1742-1745.
Moschetti, R., R. Caizzi et al., 1996. Segregation Distortion in Drosophila melanogaster: genomic organization of Rsp sequences. Genetics 144: 1665-1671.
Myers, E.W., G.G. Sutton et al., 2000. A whole-genome assembly of Drosophila. Science 287: 2196-2204.
Palopoli, M.F. & C.I. Wu, 1996. Rapid evolution of a coadapted gene complex: evidence from the Segregation Distorter (SD) system of meiotic drive in Drosophila melanogaster. Genetics 143: 1675-1688.
Palopoli, M.F., P. Doshi, et al., 1994. Characterization of two Segregation distorter revertants: evidence that the tandem duplication is necessary for Sd activity in Drosophila melanogaster. Genetics 136: 209–215.
Pimpinelli, S. & P. Dimitri, 1989. Cytogenetic analysis of Segregation Distortion in Drosophila melanogaster: the cytological organization of the Responder (Rsp) locus. Genetics 121: 765–772.
Powers, P.A. & B. Ganetzky, 1991. On the components of segregation distortion in Drosophila melanogaster. V. Molecular analysis of the Sd locus. Genetics 1991: 133–144.
Temin, R.G. & M. Marthas, 1984. Factors influencing the effect of segregation distortion in natural populations of Drosophila melanogaster. Genetics 107: 375–393.
Temin, R.G., B. Ganetzky, et al., 1991. Segregation distortion in Drosophila melanogaster: genetic and molecular analyses. Am. Nat. 137: 287–331.
Wu, C.I. & M.F. Hammer, 1991. Molecular evolution of ultraselfish genes of meiotic drive systems, in Evolution at the Molecular Level, edited by Selander, Clark & Whittam. Sinauer Associates Inc., Sunderland, MA.
Wu, C.-I., T.W. Lyttle, M.-L. Wu & G.F. Lin, 1988. Association between a satellite DNA sequence and the Responder of Segregation Distorter in D. melanogaster. Cell 54: 179–189.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Houtchens, K., Lyttle, T.W. Responder (Rsp) Alleles in the Segregation Distorter (SD) System of Meiotic Drive in Drosophila may Represent a Complex Family of Satellite Repeat Sequences. Genetica 117, 291–302 (2003). https://doi.org/10.1023/A:1022968801355
Issue Date:
DOI: https://doi.org/10.1023/A:1022968801355