Abstract
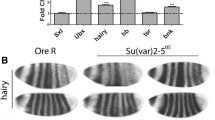
This review summarizes genetic, molecular and biochemical studies of the SU(VAR)3-9 protein and the evidence for its key role in heterochromatin formation and heterochromatic gene silencing. The Su(var)3-9 locus was first identified as a dominant modifier of position-effect variegation (PEV) in Drosophila melanogaster. Together with Su(var)2-5 and Su(var)3-7, Su(var)3-9 belongs to the group of haplo-suppressor loci which show a triplo-dependent enhancer effect. All three genes encode heterochromatin-associated proteins. Su(var)3-9 is epistatic to the PEV modifier effects of Su(var)2-5 and Su(var)3-7, and it also dominates the effect of the Y chromosome on PEV. These genetic data support a central role of the SU(VAR)3-9 protein in heterochromatic gene silencing, one that is correlated with its activity as a histone H3-K9 methyltransferase (HMTase). In fact, SU(VAR)3-9 is the main chromocenter-specific HMTase of Drosophila. SU(VAR)3-9 and HP1, the product of Su(var)2-5, are main constituents of heterochromatin protein complexes and the interaction between these two proteins is interdependent. Functional analysis in fission yeast, Drosophila and mammals demonstrate that SU(VAR)3-9-dependent gene silencing processes are conserved in these organisms. This is also demonstrated by the rescue of Drosophila Su(var)3-9 mutant phenotypes with human SUV39H1 transgenes.
Similar content being viewed by others
References
Aagaard, L., G. Laible, P. Selenko, M. Schmid, R. Dorn, G. Schotta, S. Kuhfittig, A. Wolf, A. Lebesorger, P.B. Singh, G. Reuter & T. Jenuwein, 1999. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins that complex with the heterochromatin component M31. EMBO J. 18: 1923-1938.
Appels, R. & A.J. Hilliker, 1982. The cytogenetic boundaries of the rDNA region within heterochromatin of the X chromosome of Drosophila melanogaster and their relation to male meiotic pairingsites. Genet. Res. 39: 149–156.
Bannister, A.J., P. Zegermann, J.F. Patridge, E.A. Miska, J.O. Thomas, T.C. Allshire & T. Kouzarides, 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124.
Baumbusch, L., T. Thorstensen, V. Krauss, A. Fischer, K. Naumann, R. Assalkhou, I. Schultz, G. Reuter & R.B. Aalen, 2001. The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins which can be assigned to four evolutionary conserved classes. Nucl. Acid. Res. 29: 4319-4333.
Bender, W., P. Spierer & D.S. Hogness, 1983. Chromosome walking and jumping to isolate DNA from the Ace and rosy loci and the bithorax complex of Drosophila melanogaster. J.Mol. Biol. 168: 17–33.
Boggs, B.A., P. Cheung, E. Heard, D.L. Spector, A.C. Chinault & C.D. Allis, 2002. Differential methylated forms of histone H3 show unique association patterns with inactive human X chromosome. Nat. Genet. 30: 73–76.
Cléard, F., M. Delattre & P. Spierer, 1997. SU(VAR)3-7: a Drosophila heterochromatin-associated protein and companion of HP1 in the genomic silencing of position-effect variegation. EMBO J. 16: 5280-5288.
Cléard, F. & P. Spierer, 2001. Position-effect variegation in Drosophila: the modifier Su(var)3-7 is a modular DNA-binding protein. EMBO Rep. 21: 1095-1100.
Cléard, F., M. Matsarskaia & P. Spierer, 1995. The modifier of position-effect variegation Suvar(3)7 of Drosophila: there are two alternative transcripts and seven scattered zinc fingers, each preceded by a tryptophan box. Nucl. Acid. Res. 23: 796–802.
Cooper, K.W., 1959. Cytogenetic analysis of major heterochromatic elements (especially Xh and Y) in Drosophila melanogaster, and the theory of “heterochromatin”. Chromosoma 10: 535–588.
Czermin, B., G. Schotta, B.B. Hülsmann, A. Brehm, P.B. Becker, G. Reuter & A. Imhof, 2001. Physical and functional interaction of SU(VAR)3-9 and HDAC1 in Drosophila. EMBO Rep. 2: 915–919.
Delattre, M., A. Spierer, C.-H. Tonka & P. Spierer, 2000. The genomic silencing of position-effect variegation in Drosophila melanogaster: interaction between the heterochromatinassociated proteins Su(var)3-7 and HP1. J. Cell. Sci. 113: 4253-4261.
Donaldson, K.M., A. Lui, G.H. Karpen, 2002. Modifiers of terminal deficiency-associated position effect variegation in Drosophila. Genetics 160: 995-1009.
Dorn, R., J. Szidonya, G. Korge, M. Sehnert, H. Taubert, I. Archoukieh, B. Tschiersch, H. Morawietz, G. Wustmann, G. Hoffmann & G. Reuter, 1993. P transposon-induced dominant enhancer mutations of position-effect variegation in Drosophila melanogaster. Genetics 133: 279–290.
Eissenberg, J.C., T.C. James, D.M. Foster-Hartnett, T. Hartnett, V. Ngan & S.C.R. Elgin, 1990. Mutation in a heterochromatinspecific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc. Nat. Acad. Sci. USA 87: 9923-9927.
Eissenberg, J.C. & S.C.R. Elgin, 2000. The HP1 protein family: getting a grip on chromatin. Curr. Opin. Gen. Dev. 10: 204–210.
Eissenberg, J.C., G.D. Morris, G. Reuter & T. Hartnett, 1992. The heterochromatin-associated protein HP-1 is an essential protein in Drosophila with dosage-dependent effects on position-effect variegation. Genetics 131: 345–352.
Ekwall, K. & T. Ruusala, 1994. Mutations in rik1, cir2, clr3 and clr4 genes asymmetrically derepress the silent mating-type locus in fission yeast. Genetics 136: 53–64.
Ekwall, K., E.R. Nimmo, J.P. Javerzat, B. Borgstrom, R. Egel, G. Cranston & R. Allshire, 1996. Mutations in the fission yeast silencing factors clr4+ and rik1+ disrupt the localisation of the chromo domain protein Swi6p and impair centromere function. J. Cell. Sci. 109: 2637-2648.
Fanti, L., G. Giovinazzo, M. Berloco & S. Pimpinelli, 1998. The heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol. Cell. 2: 527–538.
Finnin, M.S., J.R. Donigian, A. Cohen, V.M. Richon, R.A. Rifkind, P.A. Marks, R. Breslow & N.P. Pavletich, 1999. Structures of histone deacetylase homologue bound to TSA and SAHA. Nature 401: 188–193.
Firestein, R., X. Cui, P. Huie & M.L. Cleary, 2000. SET-domain dependent regulation of transcriptional silencing and growth control by SUV39H1, a mammalian ortholog of Drosophila Su(var)3-9. Mol. Cell. Biol. 20: 4900-4909.
Gaudin, V., M. Libault, S. Pouteau, T. Juul, G. Zhao, D. Lefebvre & O. Grandjean, 2001. Mutations in LIKE HETEROCHROMATIN PROTEIN 1 affect flowering time and plant architecture in Arabidopsis. Development 128: 4847-4858.
Hwang, K.K., J.C. Eissenberg & H.J. Worman, 2001. Transcriptional repression of euchromatic genes by Drosophila heterochromatin protein 1 and histone modifiers. Proc. Natl. Acad. Sci. USA 98: 11423-11427.
Ivanova, A.V., M.J. Bonaduce, S.V. Ivanov & A.J.S. Klar, 1998. The chromo and SET domains of the Clr4 protein are essential for silencing in fission yeast. Nat. Genet. 19: 192–195.
Jackson, J.P., A.M. Lindroth, X. Cao & S.E. Jacobson, 2002. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416: 556–560.
James, T.C. & S.C.R. Elgin, 1986. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol. Cell. Biol. 6: 3862-3872.
James, T.C., J.C. Eissenberg, C. Craig, V. Dietrich, A. Hobson & S.C.R. Elgin, 1989. Distribution patterns of HP1, a heterochromatin-associated nonhistone chromosomal protein of Drosophila. Eur. J. Cell. Biol. 50: 170–180.
Jenuwein, T., G. Laible, R. Dorn & G. Reuter, 1998. SET domain proteins modulate chromatin domains in eu-and heterochromatin. Cell. Mol. Life Sci. 54: 80–93.
Jones, R.S. & W.M. Gelbart, 1993. The Drosophila Polycombgroup gene Enhancer of zeste contains a region with seqeunce similarity to trithorax. Mol. Cell. Biol. 13: 6357-6366.
Krauss, V. & G. Reuter, 2000. Two genes become one: the genes encoding heterochromatin protein SU(VAR)3-9 and translation initiation factor subunit eIF-2γ are joined to a dicistronic unit in holometabolic insects. Genetics 156: 1157-1167.
Kuhfittig, S., J. Szabad, G. Schotta, J. Hoffmann, E. Máthé & G. Reuter, 2001. pitkin D a novel gain-of-function enhancer of position-effect variegation affects chromatin regulation during oogenesis and early embryogenesis in Drosophila. Genetics 157: 1227-1244.
Lachner, M., D. O'Carroll, S. Rea, K. Mechtler & T. Jenuwein, 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410: 116–120.
Larsson, J., J. Zhang & A. Rasmuson-Lestander, 1996. Mutations in the Drosophila melanogaster S-adenosylmethionine synthase suppress position-effect variegation. Genetics 143: 887–896.
Lefevre, G., 1976. The polytene chromosomes, pp. 31–66 in The Genetics and Biology of Drosophila, Vol. 1a, edited by M. Ashburner & E. Novitski. Academic Press, London, New York & San Francisco.
Locke, J., M.A. Kotarski & K.D. Tartof, 1988. Dosage-dependent modifiers of position-effect variegation in Drosophila and a mass action model that explains their effect. Genetics 120: 181–198.
Maison, C., D. Bailly, A.H.F.M. Peters, J.-P. Quivy, D. Roche, A. Taddei, M. Lachner, T. Jenuwein & G. Almouzni, 2002. Higherorder structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 30: 329–334.
Melcher, M., M. Schmid, L. Aagaard, P. Selenko, G. Laible & T. Jenuwein, 2000. Structure-function analysis of SUV39H1 reveals a dominant role in heterochromatin organization, chromosome segregation and mitotic progression. Mol. Cell. Biol. 20: 3728-3741.
Mottus, R., R.E. Sobels & T.A. Grigliatti, 2000. Mutational analysis of a histone deacetylase in Drosophila melanogaster: missence mutations suppress gene silencing associated with position effect variegation. Genetics 154: 657–668.
Muller, H.J., 1930. Types of visible variations induced by X-rays in Drosophila. J. Genet. 22: 299–334.
Nakayama, J., J.D. Rice, B.D. Stahl, C.D. Allis & S.I.S. Grenwal, 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292: 110–113.
O'Carroll, D., H. Scherthan, A.H. Peters, S. Opravil, A.R. Haynes, G. Laible, S. Rea, M. Schmid, A. Lebersorger, M. Jerratsch, L. Sattler, M.G. Mattei, P. Denny, S.D. Brown & T. Jenuwein, 2000. Isolation and characterization of Suvh39h2, a second histone H3 methyltransferase gene that displays testis-specific expression. Mol. Cell. Biol. 20: 9423-9433.
Peters, A.H.F.M., D. O'Carroll, H. Scherthan, K. Mechtler, S. Sauer, C. Schäfer, K. Weipoltshammer, M. Pagani, M. Lachner, A. Kohlmaier, S. Oprival, M. Doyle, M. Sibilia & T. Jenuwein, 2001. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107: 323–337.
Peters, A.H.F.M., J.E. Mermoud, D. O'Carroll, M. Pagani, D. Schweizer, N. Brockdorff & T. Jenuwein, 2002. Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin. Nat. Genet. 30: 77–80.
Raff, J.R., R. Kellum & B. Alberts, 1994. The Drosophila GAGA transcription factor is associated with specific regions of heterochromatin throughout the cell cycle. EMBO J. 13: 5977-5983.
Reuter, G. & P. Spierer, 1992. Position-effect variegation and chromatin proteins. BioEssays 14: 605–612.
Reuter, G. & J. Szidonya, 1983. Cytogenetic analysis of variegation suppressors and a dominant temperature sensitive lethal in region 23-26 of 2L in Drosophila melanogaster. Chromosoma 88: 277–285.
Reuter, G. & I. Wolff, 1981. Isolation of dominant suppressor mutations for position-effect variegation. Mol. Gen. Genet. 182: 516–519.
Reuter, G., H.J. Hoffmann & I. Wolff, 1983. Genetic study of position-effect variegation in Drosophila melanogaster: In(1)w m4h as a standard rearrangement for the isolation and characterization of suppressor and enhancer mutants. Biol. Zbl. 102: 281–298.
Reuter, G., I. Wolff & B. Friede, 1985. Functional properties of the heterochromatic sequences inducing w m4 position-effect variegation in Drosophila melanogaster. Chromosoma 93: 132–139.
Reuter, G., R. Dorn, G. Wustmann, B. Friede & G. Rauh, 1986. Third chromosome suppressor of position-effect variegation loci in Drosophila melanogaster. Mol. Gen. Genet. 202: 481–487.
Reuter, G., J. Gausz, H. Gyurkovics, B. Friede, R. Bang, A. Spierer, L.M.C. Hall & P. Spierer, 1987. Modifiers of position-effect variegation in the region from 86C to 88B of the Drosophila melanogaster third chromosome. Mol. Gen. Genet. 210: 429–436.
Reuter, G., N. Giarre, J. Farah, J. Gausz, A. Spierer & P. Spierer, 1990. Dependence of position-effect variegation in Drosophila on dose of a gene encoding an unusual zinc-finger protein. Nature 344: 219–223.
Schotta, G. & G. Reuter, 2000. Controlled expression of tagged proteins in Drosophila using a new modular P-element vector system. Mol. Gen. Genet. 262: 916–920.
Schotta, G., A. Ebert, V. Krauss, A. Fischer, J. Hoffmann, S. Rea, T. Jenuwein, R. Dorn & G. Reuter, 2002. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 21: 1121-1131.
Schultz, J., 1950. Interrelations of factors affecting heterochromatin-induced variegation in Drosophila. Genetics 35: 134.
Shareef, M.M., C. King, M. Damaj, R. Badagu, D.W. Huang & R. Kellum, 2001. Drosophila heterochromatin protein 1 (HP1)/origin recognition complex (ORC) protein is associated with HP1 and ORC and functions in heterochromatin induced silencing. Mol. Biol. Cell. 12: 1671-1685.
Szidonya, J. & G. Reuter, 1988. Cytogenetic analysis of the echinoid(ed), dumpy(dp) and clot(cl) region in Drosophila melanogaster. Genet. Res. Camb. 51: 197–208.
Tamaru, H. & E.U. Selker, 2001. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414: 277–283.
Török, T., M. Gorjanaczet, P.J. Bryant & I. Kiss, 2000. Prod is a novel DNA-binding protein that binds to the 1.686 g/cm3 10 bp satellite repeat of Drosophila melanogaster. Nucl. Acids. Res. 28: 3551-3557.
Török, T., P.D. Harvie, M. Buratovich & P.J. Bryan, 1997. The product of proliferation disrupter is concentrated at centromeres and required for mitotic chromosome condensation and cell proliferation in Drosophila. Gen. Dev. 11: 213–216.
Tschiersch, B., A. Hofmann, V. Krauss, R. Dorn, G. Korge & G. Reuter, 1994. The protein encoded by the Drosophila position effect variegation suppressor gene Su(var)3-9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J. 13: 3822-3831.
Vaute, O., E. Nicolas, L. Vandal & D. Trouche, 2002. Functional and physical interaction between the histone methyl transferase Suv39H1 and histone deacetylases. Nucl. Acid. Res. 30: 475–481.
Westphal, T. & G. Reuter, 2002. Crossing over suppression by heterochromatin and the dominant recombinogenic effects of suppressor of position-effect variegation mutations in Drosophila. Genetics 160: 609–621.
Wustmann, G., J. Szidonya, H. Taubert & G. Reuter, 1989. The genetics of position-effect modifying loci in Drosophila melanogaster. Mol. Gen. Genet. 217: 520–527.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Schotta, G., Ebert, A. & Reuter, G. SU(VAR)3-9 is a Conserved Key Function in Heterochromatic Gene Silencing. Genetica 117, 149–158 (2003). https://doi.org/10.1023/A:1022923508198
Issue Date:
DOI: https://doi.org/10.1023/A:1022923508198