Abstract
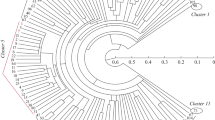
Two long primers of 19 (F17) and 20 (F13) nucleotides, respectively,were used in polymerase chain reactions to amplify DNA from differentcultivated barley accessions. These primers can distinguish closely relatedvarieties and, with a unique primer, all the barley accessions analysedshowed a characteristic fingerprint. Sixty per cent and 76% of thefragments generated using F13 and F17, respectively, were polymorphic.The genetic similarity values between accessions were estimated from F13and F17 data. The dendrogram and principal coordinate analysis performedwith F13 data revealed a clear separation of these varieties in accord withtheir pedigree relationships.
Similar content being viewed by others
References
Dellaporta, S.L., J. Wood & J.B. Hicks, 1983.A plant DNA minipreparation: Version II. Plant Mol Biol Rep 1: 19-21.
Díaz-Perales, A., R. Linacero & A.M. Vázquez, 2001. DNA ampli-fication fingerprinting using two long primers. Biotechniques 30: 718-720.
Ellis, R.P., J.W. McNicol, E. Baird, A. Booth, P. Lawrence, B. Thomas & W. Powell, 1997. The use of AFLPs to examine genetic relatedness in barley. Mol Breed 3: 359-369.
Hintum, T.J.L. van, & D. van Haalman, 1994. Pedigree analysis for composing a core collection of modern cultivars, with examples from barley (Hordeum vulgare s. lat.). Theor Appl Genet 88: 70-74.
Kalendar, R., T. Grob, M. Regina, A. Suoniemi & A. Schulman, 1999. IRAP and REMAP: two new retrotransposon-based DNA fingerprinting techniques. Theor Appl Genet 98: 704-711.
Kalendar, R., J. Tanskanen, S. Immonen, E. Nevo & A.H. Schulman, 2000. Genome evolution of wild barley (Hordeum vulgare) by BARE-1 retrotransposon dynamics in response to sharp microclimatic divergence. Proc Natl Acad Sci USA 97: 6603-6607.
Kumekawa, N., H. Ohtsubo, T. Horiuchi & E. Ohtsubo, 1999. Identification and characterization of novel retrotransposons of the gypsy type in rice. Mol Gen Genet 260: 593-602.
Linacero, R., E. Alves & A.M. Vázquez, 2000. Hot spots of DNA instability revealed through the study of somaclonal variation in rye. Theor Appl Genet 100: 506-511.
Liu, F., R. von Bothmer & B. Salomon, 2000. Genetic diversity in European accessions of the Barley Core Collection as detected by isozyme electrophoresis. Genet Res Crop Evol 47: 571-581.
Liu, F., G.L. Sun, B. Salomon & R. von Bothmer, 2001. Distribution of allozymic alleles and genetic diversity in the American Barley Core Collection. Theor Appl Genet 102: 606-615.
Melchinger, A.E., A. Graner, M. Singh & M.M. Messmer, 1994. Relationships among European barley germplasm: I. Genetic diversity among winter and spring cultivars revealed by RFLPs. Crop Sci 34: 1191-1199.
Noli, E., S. Salvi & R. Tuberosa, 1997. Comparative analysis of genetic relationships in barley based on RFLP and RAPD markers. Genome 40: 607-616.
Rohlf, F.J., 1992. Numerical Taxonomy and Multivariate Analysis System. Version 1.7. Owner manual.
Rueda, J., R. Linacero & A.M. Vázquez, 1998. Plant total DNA extraction. In: A. Karp, P.G. Isaac & D.S Ingram (Eds.), Molecular Tools for Screening Biodiversity, pp. 10-14. Chapman and Hall, London.
Russell, J.R., J.D. Fuller, M. Macaulay, B.G. Hatz, A. Jahoor, W. Powel & R.Waugh, 1997. Direct comparison of levels of genetic variation among barley accessions detected by RFLPs, AFLPs, SSRs and RAPDs. Theor Appl Genet 95: 714-722.
Schut, J.W., X. Qi, & P. Stam, 1977. Association between relationship measures on AFLP markers, pedigree data and morphological traits in barley. Theor Appl Genet 95: 1161-1168.
Sneath, P.H.A. & R.R. Sokal, 1973. Numerical Taxonomy. Freeman, San Francisco.
Tautz, D., 1989. Hypervariability of simple sequence as a general source of polymorphic DNA markers. Nucleic Acids Res 17: 6463-6471.
Waugh, R., K. McLean, A.J. Flavell, S.R. Pearce, A. Kumar, B.B.T. Thomas & W. Powell, 1997. Genetic distribution of Bare-1-like retrotransposable elements in the barley genome revealed by sequence-specific amplification polymorphisms (S-SAP). Mol Gen Genet 253: 687-694.
Williams, J.G.K., A.R. Kubelik, K.J. Livak, J.A. Rafalski & S.V. Tingey, 1990 DNA polymorphism amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 18: 6531-6535.
Zabeau, M. & P. Vos, 1993. Selective restriction fragment amplification: a general method for DNA fingerprinting. European Patent Application number 92402629.7. Publication number 0534858 A1.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Díaz-Perales, A., Linacero, R. & Vázquez, A.M. Analysis of genetic relationships among 22 European barley varieties based on two PCR markers. Euphytica 129, 53–60 (2003). https://doi.org/10.1023/A:1021598227966
Issue Date:
DOI: https://doi.org/10.1023/A:1021598227966