Abstract
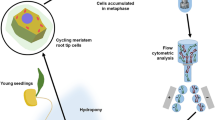
Procedures for flow cytometric analysis and sorting of mitotic chromosomes (flow cytogenetics) have been developed for chickpea (Cicer arietinum). Suspensions of intact chromosomes were prepared from root tips treated to achieve a high degree of metaphase synchrony. The optimal protocol consisted of a treatment of roots with 2 mmol/L hydroxyurea for 18 h, a 4.5-h recovery in hydroxyurea-free medium, 2 h incubation with 10 µmol/L oryzalin, and ice-water treatment overnight. This procedure resulted in an average metaphase index of 47%. Synchronized root tips were fixed in 2% formaldehyde for 20 min, and chromosome suspensions prepared by mechanical homogenization of fixed root tips. More than 4×105 morphologically intact chromosomes could be isolated from 15 root tips. Flow cytometric analysis of DAPI-stained chromosomes resulted in histograms of relative fluorescence intensity (flow karyotypes) containing eight peaks, representing individual chromosomes and/or groups of chromosomes with a similar relative DNA content. Five peaks could be assigned to individual chromosomes (A, B, C, G, H). The purity of sorted chromosome fractions was high, and chromosomes B and H could be sorted with 100% purity. PCR on flow-sorted chromosome fractions with primers for sequence-tagged microsatellite site (STMS) markers permitted assignment of the genetic linkage group LG8 to the smallest chickpea chromosome H. This study extends the number of legume species for which flow cytogenetics is available, and demonstrates the potential of flow cytogenetics for genome mapping in chickpea.
Similar content being viewed by others
References
Abbo S, Miller TE, Reader SM, Dunford RP, King LP (1994) Detection of ribosomal DNA sites in lentil and chickpea by Luorescent in situ hybridization. Genome 37: 713-716.
Arumuganathan K, Martin GB, Telenius H, Tanksley SD, Earle ED (1994) Chromosome 2-specicc DNA clones from Low-sorted chromosomes of tomato. Mol Gen Genet 242: 551-558.
Binarová P, Hause B, Doležel J, Dráber P (1998) Association of b-tubulin with kinetochore/centromeric region of plant chromosomes. Plant J 14: 751-757.
Doležel J (1991) KARYOSTAR: Microcomputer program for modelling of monoparametric Low karyotypes. Biológia 46: 1059-1064.
Doležel J, Binarová P, Lucretti S (1989) Analysis of nuclear DNA content in plant cells by Low cytometry. Biol Plant 31: 113-120.
Doležel J, Číhalíková J, Lucretti S (1992) A high-yield procedure for isolation of metaphase chromosomes from root tips of Vicia faba L. Planta 188: 93-98.
Doležel J, Lucretti S, Schubert I (1994) Plant chromosome analysis and sorting by Low cytometry. Crit Rev Plant Sci 13: 275-309.
Doležel J, Macas J, Lucretti S (1999a) Flow analysis and sorting of plant chromosomes. In: Robinson JP, Darzynkiewicz Z, Dean PN et al., eds. Current Protocols in Cytometry. New York: John Wiley & Sons, Inc., pp 5.3.1.-5.3.33.
Doležel J, Číhalíková J, Weiserová J, Lucretti S (1999b) Cell cycle synchronization in plant root meristems. Meth Cell Sci 21: 95-107.
Doležel J, Lysák MA, Kubaláková M, Šimková H, Macas J, Lucretti S (2001) Sorting of plant chromosomes. In: Darzynkiewicz Z, Crissman HA, Robinson JP, eds. Methods in Cell Biology. Vol. 64. Third Edition, Part B. San Diego: Academic Press, pp 3-31.
Fukui K, Ohmido N, Khush GS (1994) Variability in rDNA loci in the genus Oryza detected through fluorescence in situ hybridization. Theor Appl Genet 87: 893-899.
Galasso I, Pignone D (1992) Characterization of chickpea chromosomes by banding techniques. Genet Res Crop Evol 39: 115-119.
Galasso I, Frediani M, Maggiani M, Creminini R, Pignone D (1996) Chromatin organization by banding techniques, in situ hybridization, and nuclear DNA content in Cicer L. (Leguminosae). Genome 39: 258-265.
Gortner G, Nenno M, Weising K, Zink D, Nagl W, Kahl G (1998) Chromosomal localization and distribution of simple sequence repeats and the Arabidopsis-type telomere sequence in the genome of Cicer arietinum L. Chromosome Res 6: 97-104.
Gualberti G, Doležel J, Macas J, Lucretti S (1996) Preparation of pea (Pisum sativum L.) chromosome and nucleus suspensions from single root tips. Theor Appl Genet 92: 744-751.
Hüttel B, Winter P, Weising K, Choumane W, Weigand F, Kahl G (1999) Sequence-tagged microsatellite site markers for chickpea (Cicer arietinum L.). Genome 42: 210-217.
Kejnovský E, Vrána J, Matsunaga S et al. (2001) Localization of male-speciccally expressed MROS genes of Silene latifolia by PCR on Low-sorted sex chromosomes and autosomes. Genetics 158: 1269-1277.
Koblížková A, Doležel J, Macas J (1998) Subtraction with 3′ modiced oligonucleotides eliminates ampliccation artefacts in DNA libraries enriched for microsatellites. BioTechniques 25: 32-38.
Kubaláková M, Lysák MA, Vrána J, Šimková H, Číhalíková J, Doležel J (2000) Rapid identification and determination of purity of Low-sorted plant chromosomes using C-PRINS. Cytometry 41: 102-108.
Kubaláková M, Vrána J, Číhalíková J, Lysák MA, Doležel J (2001) Localisation of DNA sequences on plant chromosomes using PRINS and C-PRINS. Meth Cell Sci 23: 71-82.
Ladizinsky D (1998) Plant Evolution under Domestication. Dordrecht: Kluwer Academic Publishers, pp 254.
Lee JH, Arumuganathan K (1999) Metaphase chromosome accumulation and Low karyotypes in rice (Oryza sativa L.) root tip meristem cells. Mol Cells 9: 436-439.
Lee JH, Arumuganathan K, Kaeppler SM, Kaeppler HF, Papa CM (1996) Cell synchronization and isolation of metaphase chromosomes from maize (Zea mays L.) root tips for Low cytometric analysis and sorting. Genome 39: 697-703.
Lee JH, Arumuganathan K, Yen Y, Kaeppler S, Kaeppler H, Baenziger PS (1997) Root tip cell cycle synchronization and metaphase-chromosome isolation suitable for Low sorting in common wheat (Triticum aestivum L.). Genome 40: 633-638.
Lev-Yadun S, Gopher A, Abbo S (2000) The cradle of agriculture. Science 288: 1602-1603.
Lucretti S, Doležel J, Schubert I, Fuchs J (1993) Flow karyotyping and sorting of Vicia faba chromosomes. Theor Appl Genet 85: 665-672.
Lysák MA, Číhalíková J, Kubaláková M, Šimková H, Künzel G, Doležel J (1999) Flow karyotyping and sorting of mitotic chromosomes of barley (Hordeum vulgare L.). Chromosome Res 7: 431-444.
Macas J, Doležel J, Lucretti S et al. (1993) Localization of seed protein genes on Low-sorted celd bean chromosomes. Chromosome Res 1: 107-115.
Macas J, Gualberti G, Nouzová M, Samec P, Lucretti S, Doležel J (1996) Construction of chromosome-specific DNA libraries covering the whole genome of field bean (Vicia faba L.). Chromosome Res 4: 531-539.
Neumann P, Lysák M, Doležel J, Macas J (1998) Isolation of chromosomes from Pisum sativum L. hairy root cultures and their analysis by Low cytometry. Plant Sci 137: 205-215.
Neumann P, Požárková D, Vrána J, Doležel J, Macas J (2002) Chromosome sorting and PCR-based physical mapping in pea (Pisum sativum L.). Chromosome Res 10: 63-71.
Ocampo B, Venora G, Erico A, Singh KB, Saccardo F (1992) Karyotype analysis in the genus Cicer. J Genet Breed 46: 229-240.
Ohri D (1999) Cytology of Cicer songaricum Steph. ex DC, a wild relative of chickpea. Genet Res Crop Evol 46: 111-113.
Ohri D, Pal M (1991) The origin of chickpea (Cicer arietinum L.): karyotype and nuclear DNA content. Heredity 66: 367-372.
Požárková D, Koblížková A, Roman B et al. (2002) Development and characterization of microsatellite markers from chromosome 1-specicc DNA libraries of Vicia faba. Biol Plant 45: 337-345.
Ratnaparkhe MP, Santra DK, Tullu A, Muehlbauer FJ (1998) Inheritance of inter-simple-sequence-repeat polymorhisms and linkage with a fusarium wilt resistance gene in chickpea. Theor Appl Genet 96: 348-353.
Richards EJ, Ausubel FM (1988) Isolation of a higher eukaryotic telomere from Arabidopsis thaliana. Cell 53: 127-136.
Saxena MC (1992) Current status and prospects of Kabuli chickpea production. In: Singh KB, Saxena MC (eds.) Disease Resistance Breeding in Chickpea. Aleppo, Syria: ICARDA, pp 1-10.
Schwarzacher T, Heslop-Harrison P (2000) Practical in situ Hybridisation. Oxford, UK: BIOS Scienticc Publishers Limited, pp 203.
Simon CJ, Muehlbauer FJ (1997) Construction of a chickpea linkage map and its comparison with maps of pea and lentil. J Hered 88: 115-119.
Šimková H, Číhalíková J, Vrána J, Lysák MA, Doležel J (2003) Preparation of high molecular weight DNA from plant nuclei and chromosomes isolated from root tips. Biol Plant 46 (in press).
Tayyar RI, Lukaszewski AJ, Waines JG (1994) Chromosome banding patterns in annual species of Cicer. Genome 37: 656-663.
Ten Hoopen R, Manteuffel R, Doležel J, Malysheva L, Schubert I (2000) Evolutionary conservation of kinetochore protein sequences in plants. Chromosoma 109: 482-489.
van der Maesen LJG (1987) Origin, history, and taxonomy of chickpea. In: Saxena MC, Singh KB, eds. The Chickpea. Wallingford,UK: CAB International Publications, pp 11-34.
Vavilov NI (1926) Studies on Origin of Cultivated Plants. Leningrad: Institute of Applied Botany and Plant Breeding.
Venora G, Ocampo B, Singh KB, Saccardo F (1995) Karyotype of the kabuli-type chickpea (Cicer arietinum L.) by image analysis system. Caryologia 48: 147-155.
Vrána J, Kubaláková M, Šimková H, Číhalíková J, Lysák MA, Doležel J (2000) Flow-sorting of mitotic chromosomes in common wheat (Triticum aestivum L.). Genetics 156: 2033-2041.
Winter P, Kahl G (1995) Molecular marker technologies for plant improvement. World J Microbiol Biotechnol 11: 438-448.
Winter P, Pfaff T, Udupa SM et al. (1999) Characterization and mapping of sequence-tagged microsatellite sites in the chickpea (Cicer arietinum L.) genome. Mol Gen Genet 262: 90-101.
Winter P, Benko-Iseppon AM, Hüttel B et al. (2000) A linkage map of the chickpea (Cicer arietinum L.) genome based on recombinant inbred lines from a C. arietinum = C. reticulatum cross: localization of resistance genes for fusarium wilt races 4 and 5. Theor Appl Genet 101: 1155-1163.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Vláčilová, K., Ohri, D., Vrána, J. et al. Development of flow cytogenetics and physical genome mapping in chickpea (Cicer arietinum L.). Chromosome Res 10, 695–706 (2002). https://doi.org/10.1023/A:1021584914931
Issue Date:
DOI: https://doi.org/10.1023/A:1021584914931