Abstract
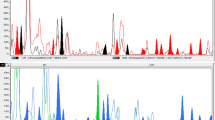
Comparative analyses of genetic diversity and phylogenetic relationshipsof sweetpotato (Ipomoea batatas (L.) Lam.) and its wildrelatives in Ipomoea series Batataswere conducted using amplified fragment length polymorphism (AFLP) and sequencedata from the internal transcribed spacer (ITS) region of the ribosomal DNA. LowITS divergence among thirteen species of ser. Batatasresulted in poorly resolved relationships. More variable AFLP characters werefound to be more efficient in characterizing genetic diversity and phylogeneticrelationships at both intra- and interspecific levels within ser.Batatas. Highly informative AFLP fingerprints of 36accessions representing 10 species of ser. Batatas weregenerated using only six primer combinations. Of the species examined,I. trifida was found to be the mostclosely related to I. batatas, whileI. ramosissima andI. umbraticola were the most distantlyrelated to I. batatas. The highlypolymorphic AFLP markers are a valuable tool in assessing genetic diversity andphylogenetic relationships of sweetpotato and its wild relatives.
Similar content being viewed by others
References
Austin D.F. 1978. The Ipomoea batatas complex I. Taxonomy. Bull. Torrey Bot. Club 105: 114–129.
Austin D.F. 1988. The taxonomy, evolution and genetic diversity of sweet potatoes and related wild species. In: Gregory P. (ed.), Exploration, Maintenance, and Utilization of Sweet Potatoes Genetic Resources. International Potato Center, Lima, Peru, pp. 27–59.
Austin D.F. 1991. Ipomoea littoralis (Convolvulaceae) taxonomy, distribution and ethnobotany. Econ. Bot. 45: 251–256.
Austin D.F., Jarret R.L. and Johnson R. 1993. Taxonomic affinities of Ipomoea gracilis R. Brown. Bull. Torrey Bot. Club 120: 49–59.
Austin D.F. and Huaman Z. 1996. A synopsis of Ipomoea (Convolvulaceae) in the Americas. Taxon 45: 3–38.
Austin D.F. and Wilkin P. 1993. Realignment of Ipomoea peruviana (Convolvulaceae). Econ. Bot. 47: 206–207.
Baldwin B.G., Sanderson M.J., Porter J.M., Wojciechowski M.F., Campbell C. and Donoghue M.J. 1995. The ITS region of nuclear ribosomal DNA. A valuable source of evidence on angiosperm phylogeny. Ann. Missouri Bot. Gard. 82: 247–277.
Bernardi G., Mouchiroud D. and Gautier C. 1988. Compositional patterns in vertebrate genomes: conservation and change in evolution. J. Mol. Evol. 28: 7–18.
Hill M., Witsenboer H., Zabeau M., Vos P., Kesseli R. and Michelmore R. 1996. PCR-based fingerprinting using AFLPs as a tool for studying genetic relationships in Lactuca spp. Theor. Appl. Genet. 93: 1202–1210.
Huang J.C. and Sun M. 1999. A modified AFLP with fluores cence-labelled primers and automated DNA sequencer detection for efficient fingerprinting analysis in plants. Biotechnology Techniques 13: 277–278.
Huang J.C. and Sun M. 2000. Genetic diversity and relationships of sweetpotato and its wild relatives in Ipomoea series Batatas (Convolvulaceae) as revealed by inter-simple sequence repeat (ISSR) and restriction analysis of chloroplast DNA. Theor. Appl. Genet. 100: 1050–1060.
Huys G., Coopman R., Janssen P. and Kersters K. 1996. High-resolution genotypic analysis of the genus Aeromonas by AFLP fingerprinting. Int. J. Syst. Bacteriol. 46: 572–580.
Janssen P., Coopman R., Huys G., Swings J., Bleeker M., Vos P. et al. 1996. Evaluation of the DNA fingerprinting method AFLP as a new tool in bacterial taxonomy. Microbiology 142: 1881–1893.
Jarret R.L., Gawel N. and Whittemore A. 1992. Phylogenetic relationships of the sweetpotato [Ipomoea batatas (L.) Lam.]. J. Amer. Soc. Hort. Sci. 117: 633–637.
Jarret R.L. and Austin D.F. 1994. Genetic diversity and systematic relationship in sweetpotato (Ipomoea batatas) (L.) Lam.) and related species as revealed by RAPD analysis. Gen. Resour. Crop Evol. 41: 165–173.
Kardolus J.P., Van-Eck H.J. and Van den Berg R.G. 1998. The potential of AFLPs in biosystematics: a first application in-Solanum taxonomy (Solanaceae). Pl. Syst. Evol. 210: 87–103.
Kumar S., Tamura K. and Nei, M. 1993. MEGA Molecular Evolutionary Genetics Analysis (user manual), V. 1.0, Institute of Molecular Evolutionary Genetics, Pennsylvania State Univ.
Liu J.S. and Schardl C.L. 1994. A conserved sequence in internal transcribed spacer 1 of plant nuclear rRNA genes. Plant Mol. Biol. 26: 775–778.
McDonald J.A. and Austin D.F. 1990. Changes and additions in Ipomoea section Batatas (Convolvulaceae). Brittonia 42: 116–120.
McDonald J.A. and Mabry T.J. 1992. Phylogenetic systematics of New World Ipomoea (Convolvulaceae) based on chloroplast DNA restriction site variation. Plant Syst. Evol. 180: 243–259.
Miller R.E., Rausher M.D. and Manos P.S. 1999. Phylogenetic systematics of Ipomoea (Convolvulaceae) based on ITS and waxy sequences. Systematic Botany 24: 209–227.
Mueller U.G., Lipari S.E. and Milgroom M.G. 1996. Amplified fragment length polymorphism (AFLP) fingerprinting of symbiotic fungi cultured by the fungus-growing ant Cyphomyrmex minutus. Mol. Ecol. 5: 119–122.
Nickrent D.L., Schuette K.P. and Starr E.M. 1994. A molecular phylogeny of Arceuthobium (Viscaceae) based on nuclear ribosomal DNA internal transcribed spacer sequences. Am. J. Bot. 81: 1149–1160.
Sharma S.K., Knox M.R. and Ellis T.H.N. 1996. AFLP analysis of the diversity and phylogeny of lens and its comparison with RAPD analysis. Theor. Appl. Genet. 93: 751–758.
Stewart C.N. and Via L.E. 1993. A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. BioTechniques 14: 748–751.
Swofford, D.L., 1999. Phylogenetic Analysis Using Parsimony, PAUP* 4.0, beta version 4.0b2. Sinauer Associates, Inc.
Vos P., Hogers R., Bleeker M., Reijans M., Van De Lee T., Hornes M. et al. 1995. AFLP:. a new technique for DNA fingerprinting. Nucleic Acids Res. 23: 4407–4414.
White T.J., Bruns T., Lee S. and Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M.A., Gelfang D.H., Sninsky J.J. and White T.J. (eds), PCR Protocols, a Guide to Methods and Applications. Academic Press, San Diego, pp. 315–322.
Xu F. and Sun M. 2001. Comparative analysis of germplasm diversity and relationships among grain amaranths and their wild relatives (Amaranthus; Amaranthaceae) using internal transcribed spacer, amplified fragment length polymorphism, and double-primer intersimple sequence repeat markers. Mol. Phylo. Evol. 21: 372–387.
Yokota Y., Kawata T., Iida Y., Kato A. and Tanifuji S. 1989. Nucleotide sequences of the 5.8S rRNA gene and internal transcribed spacer regions in carrot and broad bean ribosomal DNA. J. Mol. Evol. 29: 294–301.
Zabeau M. and Vos P. 1993. Selective restriction fragment amplification: a general method for DNA fingerprinting. European Patent Application EP 0534858.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Huang, J., Corke, H. & Sun, M. Highly polymorphic AFLP markers as a complementary tool to ITS sequences in assessing genetic diversity and phylogenetic relationships of sweetpotato (Ipomoea batatas (L.) Lam.) and its wild relatives. Genetic Resources and Crop Evolution 49, 541–550 (2002). https://doi.org/10.1023/A:1021290927362
Issue Date:
DOI: https://doi.org/10.1023/A:1021290927362