Abstract
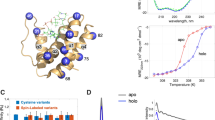
Human brain-type fatty acid-binding protein (B-FABP) has been recombinantly expressed in Escherichia coli both unlabelled and 15N-enriched for structure investigation in solution using high-resolution NMR spectroscopy. The sequential assignments of the 1H and 15N resonances were achieved by applying multidimensional homo- and heteronuclear NMR experiments. The ensemble of the 20 final energy-minimized structures, representing human B-FABP in solution, have been calculated based on a total of 2490 meaningful distance constraints. The overall B-FABP structure exhibits the typical backbone conformation described for other members of the FABP family, consisting of ten antiparallel β-strands (βA to βJ) that form two almost orthogonal β-sheets, a helix-turn-helix motif that closes the β-barrel on one side, and a short N-terminal helical loop. A comparison with the crystal structure of the same protein complexed with docosahexaenoic acid [12] reveals only minor differences in both secondary structure and overall topology. Moreover, the NMR data indicate a close structural relationship between human B-FABP and heart-type FABP with respect to fatty acid binding inside the protein cavity.
Similar content being viewed by others
References
Veerkamp JH, Maatman RGHJ: Cytoplasmic fatty acid-binding proteins: Their structure and genes. Prog Lipid Res 34: 17–52, 1995
Hohoff C, Spener F: Fatty acid binding proteins and mammaryderived growth inhibitor. Fett Lipid 100: 252–263, 1998
Veerkamp JH, Zimmerman AW: Fatty acid-binding proteins of nervous tissue. J Mol Neurosci 16: 133–142, 2001
Owada Y, Yoshimoto T, Kondo H: Spatio-temporally differential expression of genes for three members of fatty acid binding proteins in developing and mature rat brains. J Chem Neuroanat 12: 113–122, 1996
Feng L, Hatten ME, Heintz N: Brain lipid-binding protein (BLBP): A novel signaling system in the developing mammalian CNS. Neuron 12: 895–908, 1994
Kurtz A, Zimmer A, Schnütgen F, Brüning G, Spener F, Müller T: The expression pattern of a novel gene encoding brain fatty-acid binding protein correlates with neuronal and glial cell development. Development 120: 2637–2649, 1994
Godbout R, Bisgrove DA, Shkolny D, Day RS: Correlation of BFABP and GFAP expression in malignant glioma. Oncogene 16: 1955–1963, 1998
Xu LZ, Sánchez R, Sali A, Heintz N: Ligand specificity of brain lipid-binding protein. J Biol Chem 271: 24711–24719, 1996
Richieri GV, Ogata RT, Zimmerman AW, Veerkamp JH, Kleinfeld AM: Fatty acid binding proteins from different tissues show distinct patterns of fatty acid interactions. Biochemistry 39: 7197–7204, 2000
Zimmerman AW, van Moerkerk HTB, Veerkamp JH: Ligand specificity and conformational stability of human fatty acid-binding proteins. Int J Biochem Cell Biol 33: 865–876, 2001
Green P, Glozman S, Kamensky B, Yavin E: Developmental changes in rat brain membrane lipids and fatty acids: The preferential prenatal accumulation of docosahexaenoic acid. J Lipid Res 40: 960–966, 1999
Balendiran GK, Schnütgen F, Scapin G, Börchers T, Xhong N, Lim K, Godbout R, Spener F, Sacchettini JC: Crystal structure and thermodynamic analysis of human brain fatty acid-binding protein. J Biol Chem 275: 27045–27054, 2000
Zimmerman AW, Rademacher M, Rüterjans H, Lücke C, Veerkamp JH: Functional and conformational characterization of new mutants of heart fatty acid-binding protein. Biochem J 344: 495–501, 1999
Kay LE, Keifer P, Saarinen T: Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J Am Chem Soc 114: 10663–10665, 1992
Schleucher J, Sattler M, Griesinger C: Coherence selection via gradients without loss of sensitivity. The 3D-HNCO experiment. Angew Chem Int Ed Eng 32: 1489–1491, 1993
Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, Sykes BD: 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J Biomol NMR 6: 135–140, 1995
Wüthrich K: NMR of Proteins and Nucleic Acids. Wiley, New York, 1986
Pristovšek P, Lücke C, Reincke B, Ludwig B, Rüterjans H: Solution structure of the functional domain of Paracoccus denitrificans cytochrome c 552 in the reduced state. Eur J Biochem 267: 4205–4212, 2000
Güntert P, Mumenthaler C, Wüthrich K: Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol 273: 283–298, 1997
Güntert P, Braun W, Wüthrich K: Efficient computation of threedimensional protein structures in solution from nuclear magnetic resonance data using the program DIANA and the supporting programs CALIBA, HABAS and GLOMSA. J Mol Biol 217: 517–530, 1991
Wüthrich K, Billeter M, Braun W: Pseudo-structures for the 20 common amino acids for use in studies of protein conformations by measurements of intramolecular proton-proton distance constraints with nuclear magnetic resonance. J Mol Biol 169: 949–961, 1983
Dauber-Osguthorpe P, Roberts VA, Osguthorpe DJ, Wolff DJ, Genest M, Hagler AT: Structure and energetics of ligand binding to proteins: E. coli dihydrofolate reductase trimethoprin, a drugreceptor system. Proteins 4: 31–47, 1988
Laskowski RA, MacArthur MW, Moss DS, Thornton JM: AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J Appl Crystallogr 26: 283–291, 1993
Noy N: Retinoid-binding proteins: Mediators of retinoid action. Biochem J 348: 481–495, 2000
Folli C, Calderone V, Ottonello S, Bolchi A, Zanotti G, Stoppini M, Rudolfo B: Identification, retinoid binding, and X-ray analysis of a human retinol-binding protein. Proc Natl Acad Sci USA 98: 3710–3715, 2001
Lücke C, Pérez C, Cavazzini D, Rademacher M, Ludwig C, Spisni A, Rossi GL, Rüterjans H: Structure and backbone dynamics of apo-and holo-cellular retinol-binding protein in solution. J Biol Chem 277: 21983–21997, 2002
Gutiérrez-González LH, Ludwig C, Hohoff C, Rademacher M, Hanhoff T, Rüterjans H, Spener F, Lücke C: Solution structure and backbone dynamics of human epidermal-type fatty acid-binding protein (E-FABP). Biochem J 364: 725–737, 2002
Hodsdon ME, Cistola DP: Discrete backbone disorder in the nuclear magnetic resonance structure of apo intestinal fatty acidbinding protein: Implications for the mechanism of ligand entry. Biochemistry 36: 1450–1460, 1997
Zhang F, Lücke C, Baier LJ, Sacchettini JC, Hamilton JA: Solution structure of human intestinal fatty acid-binding protein: Implications for ligand entry and exit. J Biomol NMR 9: 213–228, 1997
Lücke C, Zhang F, Rüterjans H, Hamilton JH, Sacchettini JC: Flexibility is a likely determinant of binding specificity in the case of ileal lipid binding protein. Structure 4: 785–800, 1996
Lu J, Lin C-L, Tang C, Ponder JW, Kao JLF, Cistola DP, Li E: The structure and dynamics of rat apo-cellular retinol-binding protein II in solution: Comparison with the X-ray structure. J Mol Biol 286: 1179–1195, 1999
Wang L, Li Y, Abildgaard F, Markley JL, Yan H: NMR solution structure of type II human cellular retinoic acid binding protein: Implications for ligand binding. Biochemistry 37: 12727–12736, 1998
Constantine KL, Friedrichs MS, Wittekind M, Jamil H, Chu CH, Parker RA, Goldfarb V, Mueller L, Farmer BT: Backbone and side chain dynamics of uncomplexed human adipocyte and muscle fatty acid-binding proteins. Biochemistry 37: 7965–7980, 1998
Lassen D, Lücke C, Kveder M, Mesgarzadeh A, Schmidt JM, Specht B, Lezius A, Spener F, Rüterjans H: Three-dimensional structure of bovine heart fatty-acid-binding protein with bound palmitic acid, determined by multidimensional NMR spectroscopy. Eur J Biochem 230: 266–280, 1995
Lücke C, Rademacher M, Zimmerman AW, van Moerkerk HTB, Veerkamp JH, Rüterjans H: Spin-system heterogeneities indicate a selected-fit mechanism in fatty acid binding to heart-type fatty acid-binding protein (H-FABP). Biochem J 354: 259–266, 2001
Wishart DS, Sykes BD, Richards FM: Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J Mol Biol 222: 311–333, 1991
Lücke C, Huang S, Rademacher M, Rüterjans H: New insights into intracellular lipid binding proteins: The role of buried water. Prot Sci (in press)
Young ACM, Scapin G, Kromminga A, Patel SB, Veerkamp JH, Sacchettini JC: Structural studies on human muscle fatty acid binding protein at 1.4 Å resolution: Binding interactions with three C18 fatty acids. Structure 2: 523–534, 1994
Kraulis PJ: MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr 24: 946–950, 1991
Merritt EA, Bacon DJ: Raster3D: Photorealistic molecular graphics. Meth Enzymol 277: 505–524, 1997
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Rademacher, M., Zimmerman, A.W., Rüterjans, H. et al. Solution structure of fatty acid-binding protein from human brain. Mol Cell Biochem 239, 61–68 (2002). https://doi.org/10.1023/A:1020566909213
Issue Date:
DOI: https://doi.org/10.1023/A:1020566909213