Abstract
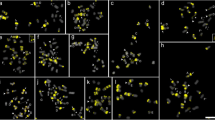
Double fluorochrome staining with chromomycin A3 (CMA) and 4′-6-diamidino-2-phenylindole (DAPI) was used to characterize and compare the distribution of constitutive heterochromatin along chromosomes of Citrus, Poncirus and Fortunella species. Only CMA-positive bands were distinguishable in metaphase chromosomes. Preferential distribution of heterochromatin in terminal regions, mainly of the long arm, and centromeric regions of a few long chromosomes was a common feature of these genera. Heteromorphism between possible homologous chromosomes was present in the majority of species. Citrus and Poncirus revealed some remarkably uniform chromosomes without any intensively fluorescing region, whereas Fortunella cultivars were differentiated by the presence of CMA bands in all chromosomes. Through measurements assisted by a computer, amounts of CMA-positive regions were shown to be highest in Fortunella. Similarities between Citrus and Poncirus suggest little heterochromatin diversification among karyotypes of these genera, whereas Fortunella, with higher amounts and more homogenous distribution of heterochromatin, is more divergent.
Similar content being viewed by others
References
Agarwal PK (1987) Karyotype of Citrus tamurana (Tan.) Chrom Inf Serv 42: 3–5.
Barrett HC, Rhodes AM (1976) A numerical taxonomic study of affinity relationships in cultivated Citrus and its close relatives. Syst Bot 1: 105–136.
Chen Q, Liang G (1989) Study on the karyotype of Poncirus. Acta Bot Yunnanica 11: 103–106 (in Chinese).
Deumling B, Greilhuber J (1982) Characterization of hetero-chromatin in different species of the Scilla siberica group (Liliaceae) by in situ hybridization of satellite DNAs and fluorochrome banding. Chromosoma 84: 535–555.
Frost HB (1938) The genetics and cytology of Citrus. Curr Sci (special no.): 24–27.
Frost HB, Soost RK (1968) Seed reproduction: development of gametes and embryos. In: Reuther W, Batchelor LD, Webber HJ, eds. The Citrus Industry. Berkeley: University of California Press, pp 290–324.
Fukui K (1986) Standardization of karyotyping plant chromosomes by a newly developed chromosome image analyzing system (CHIAS). Theor Appl Genet 72: 27–32.
Galasso I, Schmidt T, Pignone D, Heslop-Harrison JS (1995) The molecular cytogenetics of Vigna unguiculata (L.) Walp: the physical organization and characterization of 18s-5.8s-25s rRNA genes, 5s rRNA genes, telomere-like sequences, and a family of centromeric repetitive DNA sequences. Theor Appl Genet 91: 928–935.
Guerra M (1984) Cytogenetics in Rutaceae. II. Nuclear DNA content. Caryologia 37: 219–226.
Guerra M (1985) Cytogenetics of Rutaceae. III. Heterochromatin patterns. Caryologia 38: 335–346.
Guerra M (1993) Cytogenetics of Rutaceae. V. High chromosomal variability in Citrus species revealed by CMA/DAPI staining. Heredity 71: 234–241.
Handa T, Ishizawa Y, Oogaki C (1986) Phylogenetic study of fraction I protein in the genus Citrus and its close related genera. Jpn J Genet 61: 15–24.
Hirai M, Kajiura I (1987) Genetic analysis of leaf isozymes in citrus. Jpn J Breed 37: 377–388.
Ikeda H (1988) Karyomorphological studies on the genus Crepis with special reference to C-banding pattern. J Sci Hiroshima Univ, Ser B, Div 2 22: 65–117.
Ikeda F, Miranda M (1995) Aneuploidy in Eureka lemon (Citrus limon) [L.] Burm.) analyzed by image processing. J Jpn Soc Hort Sci 64(Suppl 1): 20–21 (in Japanese).
Krug CA (1943) Chromosome numbers in the subfamily Aurantioideae with special reference to the genus Citrus. Bot Gazette 104: 602–611.
Liang G (1990) Studies on the cytotaxonomy of Citrus.I. Karyotype and evolution of 30 taxa of the genera Citrus. J Wuhan Bot Res 8: 1–7 (in Chinese).
Liang GL, Yin ZY, Zeng XX, Wang Q, He H, Xu YN (1992) Analysis of karyotypes and C-banding patterns of two taxa of loose-skin oranges. J Southwest Agric Univ 14: 320–322 (in Chinese).
Lin MS, Comings DE, Alfi OS (1977) Optical studies of the interaction of 4'-6-diamidino-2-phenylindole with DNA and metaphase chromosomes. Chromosoma 60: 15–25.
Matsuyama T, Akihama T, Ito Y, Omura M, Fukui K (1996) Characterization of heterochromatic regions in 'Trovita' orange (Citrus sinensis Osbeck) chromosomes by the fluorescent staining and FISH method. Genome 39: 941–945.
Oiyama I (1981) A technique for chromosome observation in root tip cells of Citrus. Bull Fruit Tree Res Stn D 3: 1–7.
Orford SJ, Scott NS, Timmis JN (1995) A hypervariable middle repetitive DNA sequence from Citrus. Theor Appl Genet 91: 1248–1252.
ParokonnyAS, Kenton AY, Meredity L, Owens SJ, Bennet MD (1992) Genomic divergence of allopatric sibling species studies by molecular cytogenetics of their F1 hybrids. Plant J 2: 695–704.
Schwarzacher T, Schweizer D (1982) Karyotype analysis and heterochromatin differentiation with Giemsa C-banding and fluorescent counterstaining in Cephalanthera (Orchidaceae). Plant Syst Evol 141: 91–113.
Schweizer D (1976) Reverse fluorescent chromosome banding with chromomycin and DAPI. Chromosoma 58: 307–324.
Schweizer D, Loidl J (1987) A model for heterochromatin dispersion and the evolution of C-band patterns. Chrom Today 9: 61–74.
Sharma AK, Bal AK (1957) Chromosome studies in Citrus. I. Agronomia Lusitana 19: 101–126.
Soost RK (1964) Self-incompatibility in Citrus grandis (Linn.) Osbeck. Proc Am Soc Hort Sci 84: 137–140.
Stace HM, Armstrong JA, James SH (1993) Cytoevolutionary patterns in Rutaceae. Plant Syst Evol 187: 1–28.
Swingle WT (1943) The botany of Citrus and its relatives of the orange subfamily. In: Webber HJ, Batchelor LD, eds. The Citrus Industry,Vol. I. Berkeley and Los Angeles: University of California Press, pp 129–474.
Swingle WT, Reece PC (1967) The botany of Citrus and its wild relatives. In: Reuther W, Webber HJ, Batchelor LD, eds. The Citrus Industry,Vol. I. Berkeley: Universityof California Press, pp 190–430.
Tanaka T (1954) Species Problems in Citrus. Tokyo: Society for the Promotion of Science.
Tanaka T (1961) Citrologia, Semi-centennial Commemoration Papers on Citrus Studies. Osaka: Citrologia Supporting Foundation.
Verona G, Galasso I, Pignone D (1995) Quantitative hetero-chromatin determination by means of image analysis. J Genet Breed 49: 187–190.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Miranda, M., Ikeda, F., Endo, T. et al. Comparative analysis on the distribution of heterochromatin in Citrus, Poncirus and Fortunella chromosomes. Chromosome Res 5, 86–92 (1997). https://doi.org/10.1023/A:1018409922843
Issue Date:
DOI: https://doi.org/10.1023/A:1018409922843