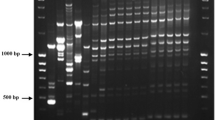
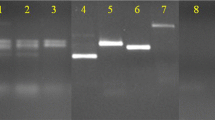
Abstract
Several polymerase chain reaction (PCR) primers were designed from the internal transcribed spacer (ITS) regions of the rDNA genes of Rosellinia necatrix to develop a PCR-based identification method. Screening the primers against two isolates of R. necatrix and six other Rosellinia species resulted in the amplification of a single specific product from R. necatrix for most of the primer pairs. Two primer pairs (R2-R8 and R10-R7) confirmed their specificity when tested against 72 isolates of R. necatrix and 93 other fungi from different hosts and geographic areas. The R10 primer was modified to obtain a Scorpion primer for detecting a specific 112bp amplicon by fluorescence emitted from a fluorophore in a self-probing PCR assay. This assay specifically recognised the target sequence of R. necatrix over a large number of other fungal species. In conventional PCR, with primer pairs R2-R8 and R10-R7, 10-fold dilutions of R. necatrix DNA indicated a detection limit of 10pgμul-1 using a single set of primers and 10fgμl-1 in nested-PCR. For Scorpion-PCR, the detection limit was 1pgμl-1 and 1fgμl-1 in nested Scorpion-PCR, i.e. 10 times more sensitive than conventional PCR. A simple and rapid procedure for DNA extraction directly from soil was modified and developed to yield DNA of purity and quality suitable for PCR assays. Combining this protocol with the nested Scorpion-PCR procedure it has been possible to specifically detect R. necatrix from artificially inoculated soils in approximately 6h.
Similar content being viewed by others
References
Bates JA, Taylor EJA, Kenyon DM and Thomas JE (2001) The application of real-time PCR to the identification, detection and quantification of Pyrenophora species in barley seed. Molecular Plant Pathology 2: 49–57
Bramwell PA, Barollon RV, Rogers HJ and Bailey MJ (1995) Extraction and PCR amplification ofDNAfrom the rhizoplane. Molecular Microbial Ecology Manual 1.4.2: 1–20
Cullen DW, Lees AK, Toth IK and Duncan JM (2001) Conventional PCR and real-time quantitative PCR detection of Helminthosporium solani in soil and potato tubers. European Journal of Plant Pathology 107: 387–398
Finetti Sialer M, Ciancio A and Gallitelli D (2000) Use of fluorogenic Scorpions for fast and sensitive detection of plant viruses. EPPO Bulletin 30: 437–440
Finetti Sialer M, Schena L and Gallitelli D (2000a) Real-Time Diagnosis in Plant pathology with self-probing amplicons (Scorpions). Proceedings of the 5th Congress of the European Foundation for Plant Pathology, Taormina, Italy, 18–22 September (pp 94–96)
Hoffman CS and Winston F (1987) A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57: 267–272
Ippolito A, Schena L, Nigro F and Salerno M (2000) Detection of Phytophthora nicotianae and P. citrophthora from roots and rhizosphere of citrus plants using PCR and Scorpion-PCR. Proceedings of the ISC 9th International Conference — Orlando, FL, USA, 3–7 December (abstract)
Lee LG, Connell CR and Bloch W (1993) Allelic discrimination by nick-translation PCR with fluorogenic probes. Nucleic Acids Research 21: 3761–3766
Lockey C, Otto E and Long Z (1998) Real-time fluorescence detection of a single DNA molecule. BioTechniques 24: 744–746
Machida U, Kami M, Fukui T, Kazuyama Y, Kinoshita M, Tanaka Y, Kanda Y, Ogawa S, Honda H and Chiba S (2000) Real-time automated PCR for early diagnosis and monitoring of cytomegalovirus infection after bone narrow transplantation. Journal of Clinical Microbiology 38: 2536–2542
Perry RL, Runkel JL and Longstroth MA (1996) The effect of rootstock on the performance of ‘Hedelfingen’ and ‘Montmorency’ cherry in Michigan, USA. Acta Horticulturae 410: 257–263
Petrini LE (1992) Rosellinia species of the temperate zones. Sydowia 44: 169–281
Reischl U, Linde H-J, Metz M, Leppmeier B and Lehn N (2000) Rapid identification of methicillin-resistant Staphylococcus aureus and simultaneous species confirmation using realtime fluorescence PCR. Journal of Clinical Microbiology 38: 2429–2433
Sambrook J, Fritsch EF and Maniatis T (1989) Molecular Cloning: A Laboratory Manual, 2nd edn, Cold Spring Harbor Laboratory Press, New York, USA
Schaad NW, Berthier-Schaad Y, Sechler A and Knorr D (1999) Detection of Clavibacter michiganensis subsp. sepedonicus in potato tubers by BIO-PCR and an automated Real time fluorescence detection system. Plant Disease 83: 1095–1100
Schena L, Finetti Sialer M and Gallitelli D (2002) Molecular detection of strain L47 of Aureobasidium pullulans, a biocontrol agent of postharvest diseases. Plant Disease 86: 54–60
Sztejnberg A and Madar Z (1980) Host range of Dematophora necatrix, the cause of white root rot disease in fruit trees. Plant Disease 64: 662–664
Sztejnberg A, Freeman S, Chet I and Katan J (1987) Control of Rosellinia necatrix in soil and in apple orchard by solarization and Trichoderma harzianum. Plant Disease 71: 365–369
Thelwell N, Millington S, Solinas A, Booth J and Brown T (2000) Mode of action and application of Scorpion primers to mutation detection. Nucleic Acids Research 28: 3752–3761
Tyagi S and Kramer FR (1996) Molecular beacons: probes that fluoresce upon hybridization. Nature Biotechnology 14: 303–308
Weller SA, Elphinstone JG, Smith NC, Boonham N and Stead DE (2000) Detection of Ralstonia solanacearum strains with a quantitative, multiplex, real-time, fluorogenic PCR (TaqMan) assay. Applied and Environmental Microbiology 66: 2853–2858
Whitcombe D, Theaker J, Guy SP, Brown T and Little S (1999) Detection of PCR products using self-probing amplicons and fluorescence. Nature Biotechnology 17: 804–807
White TJ, Bruns T, Lee S and Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ and White TJ (eds) PCR Protocols: A Guide to Methods and Applications (pp 315–322) Academic Press, Inc., New York, USA
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Schena, L., Nigro, F. & Ippolito, A. Identification and Detection of Rosellinia Necatrix by Conventional and Real-time Scorpion-PCR. European Journal of Plant Pathology 108, 355–366 (2002). https://doi.org/10.1023/A:1015697813352
Issue Date:
DOI: https://doi.org/10.1023/A:1015697813352