Abstract
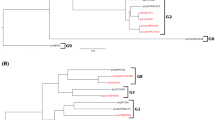
A total of 37 strains of Pseudomonas avellanae, P. syringae pv. theae and P.s. pv. actinidiae, including pathotype and reference strains, obtained from all the countries where these pathogens have been reported, were compared by means of ARDRA, repetitive PCR using ERIC, BOX and REP primer sets, whole-cell protein analysis, biochemical and nutritional tests, and pathogenicity tests. P. syringae pathovar type strains representing six genomospecies sensu Gardan et al. (1999), were also included for comparison in UPGMA cluster analysis of repetitive PCR data and SDS-PAGE of protein extracts. Among the 12 endonucleases used in ARDRA, only Tru 9I differentiated P. avellanae from P.s. pv. theae and P.s. pv. actinidiae. UPGMA cluster analysis of repetitive PCR genomic fingerprints showed 65% similarity between P.s. pv. theae and P. avellanae and 50% between the latter species and P.s. pv. actinidiae. Strains of P.s. pv. actinidiae could be grouped according to their geographic origin. Similar results were obtained with SDS-PAGE cluster analysis. PCR amplification using primers PAV 1 and PAV 22 that were developed to detect P. avellanae in apparently healthy and visibly infected hazelnut specimens yielded a band of 762bp from all strains of P. avellanae, P.s. pv. theae and P.s. pv. actinidiae. All strains lacked the syrB gene. Based on these data, we suggest that P.s. pv. actinidiae should be included in the genomospecies 8 together with P. avellanae and P.s. pv. theae. Selected biochemical and nutritional tests could differentiate these groups of strains. Pathogenicity tests clearly indicated that each group is specifically pathogenic only on the host plant species from which it was originally isolated.
Similar content being viewed by others
References
Ayers SH, Rupp P and Johnson WT (1919) A study of the alkaly-forming bacteria in milk. United States Department of Agriculture, Bulletin no. 782
Bradbury JF (1986) Guide to plant pathogenic bacteria. CAB International Mycological Institute, Kew, UK
Dice LR (1945) Measurement of the amount of ecological assiciation between species. Ecology 26: 297–302
Gardan L, Shafik H, Belouin S, Brosch R, Grimont F and Grimont PAD (1999) DNA relatedness among the pathovars of Pseudomonas syringae and description of Pseudomonas tremae sp. nov. and Pseudomonas cannabina sp. nov (ex Sutic and Dowson 1959). International Journal of Systematic Bacteriology 49: 469–478
Grifoni A, Bazzicalupo M, Di Serio C, Fancelli S and Fani R (1995) Identification of Azospirillum strains by restriction fragment length polymorphism of the 16S rDNA and the histidine operon. FEMS Microbiological Letters 127: 85–91
Janse JD, Rossi MP, Angelucci L, Scortichini M, Derks JHJ, Akkermans ADL, De Vrijer R and Psallidas PG (1996) Reclassification of Pseudomonas syringae pv. avellanae as Pseudomonas avellanae (sp. nov.), the bacterium causing canker of hazelnut (Corylus avellana L.) Systematic and Applied Microbiology 19: 589–595
Janse JD and Scortichini M (1996) Characterization of Pseudomonas syringae pv. actinidiae, the causal agent of bacterial canker of kiwifruit by whole-cell protein electrophoresis and fatty acid analysis. In: Rudolph K, Burr TJ, Mansfield JW, Stead DE, Vivian A and Von Kietzell J (eds) Pseudomonas Syringae Pathovars and Related Pathogens (p. 499) Kluwer, Dordrecht
King EO, Raney MK and Ward DE (1954) Two simple media for the demonstration of pyocianin and fluorescin. Journal of Laboratory and Clinical Medicine 44: 301–307
Lelliott RA and Stead DE (1987) Methods for the diagnosis of bacterial diseases of plants. Blackwell Scientific Publications for the British Society of Plant Pathology, Oxford
Louws FJ, Fulbright DW, Stephens CT and De Bruijn FJ (1994) Specific genomic fingerprinting of phytopathogenic Xanthomonas and Pseudomonas pathovars and strains generated with repetitive sequence and PCR. Applied and Environmental Microbiology 60: 2285–2296
Luisetti J, Prunier JP and Gardan L (1972) Un milieu pour la mise enévidence de la production d'un pigment fluorescent par Pseudomonas mors-prunorum f. sp. persicae. Annales de Phytopathologie 4: 295–296
Marques AS dosA, Corbiére R, Gardan L, Tourte C, Manceau C, Taylor JD and Samson R (2000) Multiphasic approach for the identification of the different classification levels of Pseudomonas savastanoi pv. phaseolicola. European Journal of Plant Pathology 106: 715–734
Palleroni NJ (1984) Pseudomonas Migula 1894. In: Krieg NR and Holt JG (eds) Bergey's Manual of Systematic Bacteriology (pp 141–199) Williams and Wilkins, Baltimore
Picard C, Di Cello F, Ventura M, Fani R and Guckert A (2000) Frequency and biodiversity of 2,4-diacetylphloroglucinolproducing bacteria isolated from the maize rhizosphere at different stage of plant growth. Applied and Environmental Microbiology 66: 984–955
Psallidas PG and Panagopoulos CG (1979) A bacterial canker of hazelnut in Greece caused by Pseudomonas syringae pv. avellanae. Phytopathologische Zeitschrift 94: 103–111
Psallidas PG (1993) Pseudomonas syringae pv. avellanae pathovar nov., the bacterium causing canker disease on Corylus avellana. Plant Pathology 42: 358–363
Sawada H, Suzuki F, Matsuda I and Saitou N (1999) Phylogenetic analysis of Pseudomonas syringae pathovars suggests the horizontal gene transfer of argK and the evolutionary stability of hrp gene cluster. Journal of Molecular Evolution 49: 627–644
Scortichini M (1994) Occurrence of Pseudomonas syringae pv. actinidiae in Italy. Plant Pathology 43: 1035–1038
Scortichini M and Tropiano FG (1994) Severe outbreak of Pseudomonas syringae pv. avellanae in Italy. Journal of Phytopathology 140: 65–70
Scortichini M and Lazzari M (1996) Systemic migration of Pseudomonas syringae pv. avellanae in twigs and young trees of hazelnut and symptom development. Journal of Phytopathology 144: 215–219
Scortichini M, Dettori MT, Marchesi U, Palombi MA and Rossi MP (1998) Differentiation of Pseudomonas avellanae strains from Greece and Italy by rep-PCR genomic finger-printing. Journal of Phytopathology 146: 417–420
Scortichini M and Angelucci L (1999) Phenotypic characterization of Pseudomonas avellanae (Psallidas) Janse et al., and occurrence of colony variants. Journal of Plant Pathology 81: 55–61
Scortichini M and Marchesi U (2001) Sensitive and specific detection of Pseudomonas avellanae using primers based on 16S rRNA gene sequences. Journal of Phytopathology 149: 527–532
Serizawa S and Ichikawa T (1993) Epidemiology of bacterial canker of kiwifruit. 2 The most suitable times and environments for infection on new canes. Annals of the Phytopathological Society of Japan 59: 460–468
Smith JJ, Offord LC, Holderness H and Saddler GS (1995) Genetic diversity of Burkholderia solanacearum (synonim Pseudomonas solanacearum) race 3 in Kenya. Applied and Environmental Microbiology 61: 4263–4268
Sorensen KN, Kim K-H and Takemoto JY (1998) PCR detection of cyclic lipodepsinonapeptide-producing Pseudomonas syringae pv. syringae and similarity of strains. Applied and Environmental Microbiology 64: 226–230
Takikawa Y, Ando Y, Hamaya E, Tsuyumu S and Goto M (1988) Identification of the pathogen responsible for bacteriosis of tea plant occurred in 1983. Annals of the Phytopathological Society of Japan 54: 224–228
Takikawa Y, Serizawa S, Ichikawa, Tsuyumu S and Goto M (1989) Pseudomonas syringae pv. actinidiae pv. nov.: the causal bacterium of canker of kiwifruit in Japan. Annals of the Phytopathological Society of Japan 55: 437–444
Vaneechoutte M, Rossau R, De Vos P, Gillis M, Janssens D, Paepe N, De Rouck A, Fiers T, Claeys G and Kersters K (1992) Rapid identification of bacteria of the Comamonadaceae with amplified ribosomal DNA-restriction analysis (ARDRA). FEMS Microbiological Letters 93: 227–234
Author information
Authors and Affiliations
Additional information
The author is staff member of the Istituto Sperimentale per la Patologia Vegetale, Roma, Italy temporarily assigned to ISF.
Rights and permissions
About this article
Cite this article
Scortichini, M., Marchesi, U. & Di Prospero, P. Genetic Relatedness among Pseudomonas avellanae, P. Syringae pv. Theae and P.s. pv. Actinidiae, and their Identification. European Journal of Plant Pathology 108, 269–278 (2002). https://doi.org/10.1023/A:1015178104513
Issue Date:
DOI: https://doi.org/10.1023/A:1015178104513