Abstract
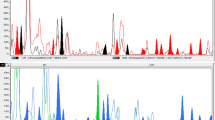
Five pathotypes belonging to formae speciales larici-epitea typica (LET), larici-retusae (LR) and larici-daphnoides (LD) of Melampsora larici-epitea were examined using amplified fragment length polymorphism (AFLP). Of 213 AFLP markers scored, several were found to be exclusive to different formae speciales. The dendrogram placed the five pathotypes into distinct groups. Within pathotypes, average Nei & Li's similarity coefficients were calculated as 0.71–0.85. The similarities were 0.66–0.72 among the three pathotypes within LET and 0.34–0.44 between pathotypes belonging to different formae speciales. When assessed using the Shannon index, the diversity within locations was estimated as 0.55–0.59, greater than that found within pathotypes (0.24–0.42). The average per-locus diversity was 0.37 among the pathotypes and 0.11 among the locations. When the data from both LET and LR isolates were examined using AMOVA, the majority of the variation (70.85%) was attributed to among pathotypes within location. When only LET types were included, approximately half of the variation was partitioned to among pathotypes within location and the other half to among the isolates within collection. It appears that the degree of differentiation of LET4 on S. × mollissima between Loughgall and Long Ashton sites has decreased markedly since 1992, when it was first detected.
Similar content being viewed by others
References
Bean WJ (1973) Trees and Shrubs Hardy in the British Isles. Vol II, 8th edn John Murray Ltd., London, UK
Bean WJ (1980) Trees and Shrubs Hardy in the British Isles. Vol IV, 8th edn John Murray Ltd., London, UK
Bussell JD (1999) The distribution of random amplified polymorphic DNA (RAPD) diversity amongst populations of Isotoma petraea (Lobeliaceae). Molecular Ecology 8: 775–789
Excoffier L, Smouse PE and Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131: 179–191
Gäumann E (1959) Die Rostpilze Mitteleuropas. Beiträge zur Kryptogamenflora Schweiz 12, Buchdruckerei Büchler & Co, Bern, Schweiz
Hiratsuka N (1932) Inoculation experiments with some heteroecious species of the Melampsoraceae in Japan. Japanese Journal of Botany 6: 1–33
Hiratsuka N and Kaneko S (1982) A taxonomic revision of Melampsora on willows in Japan. Reports of Tottori Mycological Institute 20: 1–32
Meikle RD (1984) Willows and Poplars of Great Britain and Ireland. BSBI Handbook 4. Botanical Society of British Isles, London, UK
Nei M and Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proceedings of the National Academy of Sciences of the USA 76: 5269–5273
Pei MH, Royle DJ and Hunter T (1993) Identity and host alternation of some willow rusts (Melampsora spp.) in England. Mycological Research 97: 845–851
Pei MH, Royle DJ and Hunter T (1996) Pathogenic specialization of Melampsora epitea var. epitea on Salix. Plant Pathology 45: 679–690
Pei MH, Hunter T and Ruiz C (1999a) Occurrence of Melampsora rusts in biomass willow plantations for renewable energy in the United Kingdom. Biomass and Bioenergy 17: 153–163
Pei MH, Hunter T and Royle DJ (1999b) Host—pathogen relationship between Salix and Melampsora sheds light on the parentage of some willow hybrids for biomass. New Phytologist 141: 155–160
Pei MH, Royle DJ and Hunter T (1999c) Hybridisation in larchalternating Melampsora epitea (M. larici-epitea). Mycological Research 103: 1440–1446
Pei MH and Ruiz C (2000) AFLP evidence of distinct patterns of life-cycle in two forms of Melampsora rust on Salix viminalis. Mycological Research 104: 937–942
Pei MH, Yuan ZW, Hunter T and Ruiz C (2000) Heterogeneous nature of a ‘new’ pathotype of Melampsora rust on Salix revealed by AFLP. European Journal of Plant Pathology 106: 771–779
Samils B, Lagercrantz U, Lascoux M and Gullberg U (2001) Genetic structure of Melampsora epitea populations in Swedish Salix viminalis plantations. European Journal of Plant Pathology 107: 399–409
Sneath PHA and Sokal RR (1973) Numerical Taxonomy, (573 pp) W.H. Freeman and Company, San Fransisco, USA
Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M and Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acid Research 23: 4407–4414
Sydow P and Sydow H (1915) Monographia Uredinearum 3. Bornträger, Leipzig, Germany
Yap IV and Nelson RJ (1996) WinBoot: A program for performing bootstrap analysis of binary data to determine the confidence limits of UPGMA-based dendrograms. International Rice Research Instititute, http://www.irri.org/ winboot.htm
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Pei, M., Bayon, C., Ruiz, C. et al. Genetic Variation in Melampsora Larici-epitea on Biomass willows Assessed using AFLP. European Journal of Plant Pathology 108, 229–236 (2002). https://doi.org/10.1023/A:1015126016885
Issue Date:
DOI: https://doi.org/10.1023/A:1015126016885