Abstract
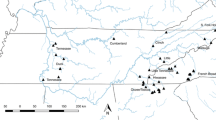
This study examines geneticstructure of central and eastern North American Loggerhead Shrike (Lanius ludovicianus)populations. Samples derived from 27populations (n = 206) covering the range of threerecognized subspecies: L. l. migrans,L. l. ludovicianus, and L. l.excubitorides, and included locales from aputative intergrade zone between twosubspecies. For L. l. migrans, samplesinclude both those from extant populations andfrom museum specimens spanning approximately130 years. For all samples we obtained 267 basepairs of mitochondrial control region sequenceand identified a total of 23 distincthaplotypes. For a subset of samplesrepresenting a locale centered on the Canadianportion of an intergrade zone between migrans and excubitorides and locales onalternate sides of the intergrade, we obtainedadditional sequence for an intron of thenuclear glyceraldehyde-3-phosphatedehydrogenase gene. Analyses of temporal trendsin control region diversity of L. l.migrans populations indicate a diminutioncoincident with the decline in populationnumbers. Results from an Analysis of MolecularVariance on the control region data showed thata significant amount of total control regionspatial variation was apportioned among thethree subspecies (24.42%; P < 0.01). Furtheranalyses indicated statistically significantdifferences between eastern and westernpopulations of L. l. migrans. We found nosignificant differences among consideredpopulations in frequencies of six identifiedGpdh alleles, although this result ispreliminary because of small sample sizes.However, both the mitochondrial and intron datasuggest a higher genetic diversity in theintergrade zone. Based on our analyses wedefine four management units for eastern andcentral populations of Loggerhead Shrikes.
Similar content being viewed by others
References
Avise JC (1994) Molecular Markers, Natural History and Evolution. Chapman and Hall, New York.
Baker C, Medrano-Gonzalez L, Calambokidis J, Perry A, Pichler F, Rosenbaum H, Straley JM, Urban-Ramirez J, Yamaguchi M, Von Ziegesar O (1998) Population structure of nuclear and mitochondrial DNA variation among Humpback Whales in the North Pacific. Mol. Ecol., 7, 695–707.
Ball RM, Freeman S, James FC, Bermingham E, Avise JC (1988) Phylogeographic population structure of Red-winged Blackbirds assessed by mitochondrial DNA. Proc. Nat. Acad. Sci. USA, 85, 1558–1562.
Ball RM, Avise JC (1992) Mitochondrial DNA phylogeographic differentiation among avian populations and the evolutionary significance of subspecies. Auk, 109, 626–636.
Boyce WM, Ramey RR, Rodwell TC, Rubin ES, Singer RS (1999) Population subdivision among Desert Bighorn Sheep (Ovis canadensis) ewes revealed by mitochondrial DNA analysis. Mol. Ecol., 8, 99–106.
Brooks BL, Temple SA (1990) Dynamics of a Loggerhead Shrike population in Minnesota. Wilson Bull., 102, 441–450.
Brown WM, Prager EM, Wang A, Wilson AC (1982) Mitochondrial DNA sequences of primates: Tempo and mode of evolution. J. Mol. Evol., 18, 225–239.
Cade TJ, Woods CP (1997) Changes in distribution and abundance of the Loggerhead Shrike. Cons. Biol., 11, 21–31.
Cadman MD (1985) Status Report on the Loggerhead Shrike (Lanius ludovicianus) in Canada. Unpublished report to the Committee on the Status of Endangered Wildlife in Canada
Cadman MD (1990) Updated Status Report on the Loggerhead Shrike (Eastern Populations) Lanius ludovicianus Migrans in Canada. Unpublished report to the Committee on the Status of Endangered Wildlife in Canada (COSEWIC). 17 pp.
Collister DM, De Smet K (1997) Breeding and natal dispersal in the Loggerhead Shrike. J. Field Ornithol., 68, 273–282.
COSEWIC (1995) Canadian Species at Risk. Committee on the Status of Endangered Wildlife in Canada. Unpublished report. 16 pp.
Cuddy D (1995) Protection and restoration of breeding habitat for the Loggerhead Shrike (Lanius ludovicianus) in Ontario, Canada. Proc. Western Fdn. Vert. Zool., 6, 283–291.
Edwards SV (1993) Mitochondrial gene genealogy and gene flow among island and mainland populations of a sedentary songbird, the Grey-crowned Babbler (Pomatostomus temporalis). Evolution, 47, 1118–1137.
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics, 131, 479–491.
Fisher RA (1930) The Genetical Theory of Natural Selection. Clarendon Press, Oxford.
Friesen VL, Congdon BC, Walsh HE, Birt TP (1997) Intron variation in Marbled Murrelets detected using analyses of singlestranded conformational polymorphisms. Mol. Ecol., 6, 1047–1058.
Fry A, Zink RM (1998) Geographic analysis of nucleotide diversity and song sparrow (Aves: Emberizidae) population history. Mol. Ecol., 7, 1303–1313.
Haas CA, Sloane SA (1989) Low return rates of migratory Loggerhead Shrikes: Winter mortality or low site fidelity? Wilson Bull., 101, 458–460.
Haig SM, Avise JC (1996) Avian conservation genetics. In: Conservation Genetics: Case Studies from Nature (eds. Avise JC, Hamrick JL) pp. 160–189. Chapman Hall, New York.
Hewitt GM (1996) Some genetic consequences of ice ages, and their role in divergence and speciation. Biol. J. Linn. Soc., 58, 247–276.
Kidd MG, Friesen VL (1998) Analysis of mechanisms of microevolutionary change in Cepphus guillemots using patterns of control region variation. Evolution, 52, 1158–1168.
Kimura M(1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol., 16, 111–120.
Kocher TD, Thomas WK, Meyer A, Edwards SV, Pääbo S, Villablanca FX, Wilson AC (1989) Dynamics of mitochondrial DNA evolution in animals: Amplification and sequencing with conserved primers. Proc. Nat. Acad. Sci. USA, 86, 6196–6200.
Lefranc N (1997) Shrikes. A Guide to the Shrikes of the World. Yale Univ. Press. New Haven, Connecticut.
Lucchini V, Randi E (1998) Mitochondrial DNA sequence variation and phylogeographical structure of Rock Partridge (Aletoris graeca) populations. Heredity, 81, 528–536.
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res., 27, 209–220.
Marshall HD, Baker AJ (1997) Structural conservation and variation in the mitochondrial control region of fringilline finches (Fringilla spp.) and the Greenfinch (Carduelis chloris). Mol. Biol. Evol., 14, 173–184.
Miller AH (1931) Systematic revision and natural history of the American shrikes (Lanius). Univ. California Publ. Zool., 38, 11–242.
Milligan BG, Leebans-Mack J, Strand AE (1994) Conservation genetics: Beyond the maintenance of marker diversity. Mol. Ecol., 3, 423–435.
Moore WS, Graham JH, Price JT (1991) Mitochondrial DNA variation in the Northern Flicker (Colaptes aurates, Aves). Mol. Biol. Evol., 8, 327–344.
Morrison ML (1981) Population trends of the Loggerhead Shrike in the United States. Am. Birds, 35, 754–757.
Moritz C (1994) Applications of mitochondrial DNA analysis in conservation: A critical review. Mol. Ecol., 3, 401–411.
Mundy NI, Unitt P, Woodruff DS (1997a) Skin from feet of museum specimens as a non-destructive source of DNA for avian genotyping. Auk, 114, 126–129.
Mundy NI, Winchell CS, Burr T, Woodruff DS (1997b) Microsatellite variation and microevolution in the critically endangered San Clemente Island Loggerhead Shrike (Lanius ludovicianus mearnsi). Proc. Roy. Soc. Lond. B., 264, 869–875.
Mundy NI, Winchell CS, Woodruff DS (1997c) Genetic differences between the endangered San Clemente Island Loggerhead Shrike Lanius ludovicianus mearnsi and two neighbouring subspecies demonstrated by mtDNA control region and cytochrome b sequence variation. Mol. Ecol., 6, 29–37.
Nagata J, Masuda R, Kaji K, Kaneko M, Yoshida MC (1998) Genetic variation and population structure of the Japanese Sika Deer (Cervus nippon) in Hokkaido Island, based on mitochondrial D-loop sequences. Mol. Ecol., 7, 871–877.
Nei M (1987) Molecular Evolutionary Genetics. Columbia University Press, New York.
Peterjohn BG, Sauer JR (1995) Population trends of the Loggerhead Shrike from the North American Breeding Bird Survey. Proc. Western Fdn. Vert. Zool. 6, 117–121.
Prescott DRC, Collister DM (1993) Characteristics of occupied and unoccupied Loggerhead Shrike territories in southeastern Alberta. J. Wildl. Manag., 57, 346–352.
Questiau S, Eybert MC, Gaginskaya AR, Gielly L, Taberlet P (1998) Recent divergence between two morphologically differentiated subspecies of Bluethroat (Aves: Muscicapidae: Luscinia svecica) inferred from mitochondrial DNA sequence variation. Mol. Ecol., 7, 239–245.
Quinn TW (1992) The genetic legacy of Mother Goose – phylogeographic patterns of Lesser Snow Goose Chen caerulescens caerulescens maternal lineages. Mol. Ecol., 1, 105–117.
Raymond M, Roussett F (1995) GENEPOP (version 3): Population genetics software for exact test and ecumenicism. J. Hered., 86, 248–249.
Rice WR (1989) Analyzing tables of statistical tests. Evolution 43, 223–225.
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, 2nd ed. Cold Spring Harbor Laboratory Press, New York.
Sauer J, Schwartz S, Hoover B (1996) The Christmas Bird Count Home Page, Version 9.5.1 Patuxent Wildlife Research Center, Laurel, MD.
Schneider S, Kueffer JM, Roessli D, Excoffier L (1997) Arlequin ver. 1.1: A software for population genetic data analysis. Genetics and Biometry Laboratory, University of Geneva, Switzerland.
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics, 123, 585–595.
Thomas WK, Pääbo S, Villablanca FX, Wilson AC (1990) Spatial and temporal continuity of kangaroo rat populations shown by sequencing mitochondrial DNA from museum specimens. J. Mol. Evol., 31, 101–112.
Van Wagner CE, Baker AJ (1990) Association between mitochondrial DNA and morphological evolution in Canada Geese. J. Mol. Evol., 31, 373–382.
Wakeley J (1993) Substitution rate variation among sites in hypervariable region I of human mitochondrial DNA. J. Mol. Evol., 37, 613–623.
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution, 38, 1358–1370.
Wenink PW, Baker AJ, Rosner HU, Tilanus MGJ (1996) Global mitochondrial DNA phylogeography of holarctic breeding Dunlins (Calidris alpina). Evolution, 50, 318–330.
Wilson AC, Cann RL, Carr SM, George M, Gyllensten UB, Helm-Bychowski KM, Higuchi RG, Palumbi SR, Prager EM, Sage RD, Stoneking m (1985) Mitochondrial DNA and two perspectives on evolutionary genetics. Biol. J. Linn. Soc., 26, 375–400.
Woodruff DS (1979) Postmating reproductive isolation in Pseudophryne and the evolutionary significance of hybrid zones. Science, 203, 561–563.
Wright S (1943) Isolation by distance. Genetics, 28, 114–138.
Wright S (1951) The genetical structure of populations. Annu. Rev. Eugen., 5, 323–354.
Zink RM (1994) The geography of mitochondrial DNA variation, population structure, hybridization, and species limits in the Fox Sparrow (Passerella iliaca). Evolution, 48, 96–111.
Zink RM (1996) Comparative phylogeography in North American birds. Evolution, 50, 308–317.
Zink RM, Dittmann DL (1993a) Gene flow, refugia, and evolution of geographic variation in the song sparrow (Melospiza melodia). Evolution, 47, 717–729.
Zink RM, Dittmann DL (1993b) Population structure and gene flow in the chipping sparrow and a hypothesis for evolution in the genus Spizella. Wilson Bull., 105, 399–413.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Vallianatos, M., Lougheed, S.C. & Boag, P.T. Conservation genetics of the loggerhead shrike (Lanius ludovicianus) in central and eastern North America. Conservation Genetics 3, 1–13 (2002). https://doi.org/10.1023/A:1014232326576
Issue Date:
DOI: https://doi.org/10.1023/A:1014232326576