Abstract
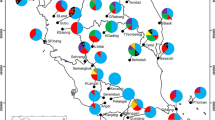
Allozyme polymorphism at seven loci (TPI, G6PD-2,IDH-1, SKD-2, MDH-1, GOT-1, andGOT-2) was employed to detect the level of geneticdiversity in C.alismatifolia populations from both cultivatedand wild habitats in Thailand. High diversity was observed in allpopulations with relatively lower values in cultivated populations.Percentage of polymorphic loci (P)varied from 85.7–100% in cultivated populations comparedwith 100% in all natural populations. Allele number per locus(A L) was 3.14 in cultivatedpopulations, and from 2.86–4 in natural populations. Allelenumber per polymorphic locus(A P) of cultivated andnatural populations ranged from 3.14–3.5 and 2.86–4,respectively. Genetic diversity within populations(H S) varied from0.586–0.611 in cultivated and from 0.621–0.653 in naturalpopulations. The genetic identity(I SP) for the species was0.833. The cultivated populations yielded higher value of geneticidentity with highland populations(I C /H =0.776) than with the lowland ones(I C /L =0.754). The analysis of genetic similarities with theNeighbor-Joining algorithm results in the separation ofcultivated populations from all wild populations. One highlandpopulation from the tourist spot, H2, was placed in a separatecluster between the cultivated and other wild populations. It isconsidered as the possible origin of the cultivatedpopulations.
Similar content being viewed by others
References
Aradhya M.K., Zee F.T. and Manshardt R.M. 1996. Genetic diversity in Nephelium as revealed by isozyme polymorphism. J. Hort. Sci. 71: 847–857.
Barrett S.C.H. and Shore J.S. 1989. Isozyme variation in colonizing plants. In: Soltis D.E. and Soltis P.S. (eds), Isozyme in Plant Biology. Dioscorides Press, Portland, Oregon, pp. 106–126.
Bretting P.K. and Widrlechner M.P. 1995. Genetic markers and horticultural germplasm management. HortScience 30: 1349–1356.
Crawford D.J. 1990. Plant Molecular Systematics. John Wiley and Sons, New York.
Doebley J.F. 1989. Isozymic evidence and evolution of crop plants. In: Soltis D.E. and Soltis P.S. (eds), Isozymes in Plant Biology. Dioscorides Press, Oregon, pp. 165–191.
Doebley J.F., Goodman M.M. and Stuber C.W. 1984. Isozymatic variation in Zea (Gramineae). Syst. Bot. 9: 203–218.
Doebley J.F., Goodman M.M. and Stuber C.W. 1985. Isozymic variation in the races of maize from Mexico. Amer. J. Bot. 72: 629–639.
Ellstrand N.C. and Marshall D.L. 1985. The impact of domestication on distribution of allozyme variation within and among cultivars of radish, Raphanus sativus L. Theor. Appl. Genet. 69: 393–398.
Ellstrand N.C. and Lee J.L. 1987. Cultivar identification of cherimoya (Annona cherimola Mill.) using isozyme markers. Sci. Hort. 32: 25–31.
Gagnepain F. 1908. Zingiberaceae. In: Lecompte H (ed.), Flore Generale de l'Indo-Chine, Tome 6 fasc. 1., pp. 25–121.
Gottlieb L.D. 1977. Electrophoretic evidence and plant systematics. Ann. Missouri Bot. Gard. 64: 161–180.
Gottlieb L.D. 1981. Electrophoretic evidence and plant populations. Progr. Phytochem. 7: 1–46.
Hamrick J.L. 1987. Gene flow and distribution of genetic variation in plant populations. In: Urbanska K.M (ed.), Differentiation Patterns in Higher Plants. Academic Press, Florida, pp. 53–67.
Hillis D.M. 1984. Misuse and modification of Nei's genetic distance. Syst. Zool. 33: 238–240.
Hunter R.L. and Markert C.L. 1957. Histochemical demonstration of enzymes separated by zone electrophoresis in starch gels. Science 125: 1294–1295.
Lewontin R.C. and Hubby J.L. 1966. A molecular approach to the study of genetic heterozygosity in natural populations. II. Amount of variation and degree of heterozygosity in natural populations of Drosophila pseudoobscura. Genetics 54: 595–609.
Loveless M.D. and Hamrick J.L. 1984. Ecological determinants of genetic structure in plant populations. Annue. Rev. Ecol. Syst. 15: 65–95.
Meerts P., Baya T. and Lefebvre C. 1998. Allozyme variation in the annual weed species complex Polygonum aviculare (Polygonaceae) in relation to ploidy level and colonizing ability. Pl. Syst. Evol. 211: 239–256.
Morishima H., Shimamoto Y., Sato Y.I., Chitrakon S., Sano Y., Barbier P. et al. 1996. Monitoring wild rice populations in permanent study sites in Thailand IRRI, Rice Genetics III., pp. 377–380.
Nei M. 1972. Genetic distance between populations. Am. Naturalist 106: 283–293.
Nei M. 1978. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89: 583–590.
Obara-Okeyo P., Fujii K. and Kako S. 1997. Enzyme polymor-phism in Cymbidium orchid cultivars and inheritance of leucine aminopeptidase. HortScience 32: 1267–1271.
Perry L.M. and Metzger J. 1980. Medicinal Plants of East and Southeast Asia. MIT Press, London.
Rohlf F.J. 1997. NTSYSpc: Numerical Taxonomy and Multivariate Analysis System, version 2.00. Exeter Software, New York.
Saitou N. and Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425.
Werth C.R. 1985. Implementing an isozyme laboratory at a field station. Virginia J. Sci. 36: 53–76.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Paisooksantivatana, Y., Kako, S. & Seko, H. Genetic diversity of Curcuma alismatifolia Gagnep.(Zingiberaceae) in Thailand as revealed by allozymepolymorphism. Genetic Resources and Crop Evolution 48, 459–465 (2001). https://doi.org/10.1023/A:1012078728003
Issue Date:
DOI: https://doi.org/10.1023/A:1012078728003