Abstract
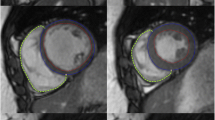
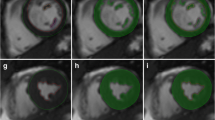
The goal of this research was to develop an automated algorithm for tracking the borders of the left ventricle (LV) in a cine-MRI gradient-echo temporal data set. The algorithm was validated on four patient populations: healthy volunteers and patients with dilated cardiomyopathy (DCM), left ventricular hypertrophy (LVH), or left ventricular aneurysm (LVA). A full tomographic set (∼11 slices/case) of short-axis images through systole was obtained for each patient. Initial endocardial and epicardial contours for the end-diastolic (ED) and end-systolic (ES) frames were manually traced on the computer by an experienced radiologist. The ED tracings were used as the starting point for the algorithm. The borders were tracked through each phase of the temporal data set, until the ES frame was reached (∼7 phases/slice). Peak gradients along equally spaced chords calculated perpendicular to a centerline determined midway between the endocardial and epicardial borders were used for border detection. This approach was tested by comparing the LV epicardial and endocardial volumes calculated at ES to those based on the manual tracings. The results of the algorithm compared favorably with both the endocardial (r 2 = 0.72 − 0.98) and epicardial (r 2 = 0.96 − 0.99) volumes of the tracer.
Similar content being viewed by others
References
Balzer P, Furber A, Cavaro-Ménard C, et al. Simultaneous and correlated detection of endocardial and epicardial borders on short-axis MR images for the measurement of left ventricular mass. RadioGraphics 1998; 18: 1009-1018.
Chakraborty A, Staib LH, Duncan JS. Deformable boundary finding in medical images by integrating gradient and region information. IEEE Transac Med Imaging 1996; 15: 859-870.
Fleagle SR, Thedens DR, Ehrhardt JC, et al. Automated identification of left ventricular borders from spin-echo magnetic resonance images. Invest Radiol 1991; 25(4): 295-303.
Furber A, Balzer P, Cavaro-Ménard C, et al. Experimental validation of an automated edge-detection method for a simultaneous determination of the endocardial and epicardial borders in short-axis cardiac MR images: application in normal volunteers. JMRI 1998; 8: 1006-1014.
Kass M, Witkin A, Terzopoulos D. Snakes: active contour models. Int J Computer Vision 1987; 1: 321-331.
Lalande A, Legrand L, Walker PM, et al. Automatic detection of left ventricular contours from cardiac cine magnetic resonance imaging using fuzzy logic. Invest Radiol 1999; 34(3): 211-217.
Lobregt S, Viergever MA. A discrete dynamic contour model. IEEE Transac Med Imaging 1995; 14(1): 12-23.
Philip KP, Dove EL, McPherson DD, et al. Automatic detection of myocardial contours in cine-computed tomographic images. IEEE Transac Med Imaging 1994; 13(2): 241-253.
van der Geest RJ, Jansen E, Buller VGM, Reiber JHC. Automated detection of left ventricular epi-and endocardial contours in short-axis MR images. Computers in Cardiology. IEEE Computer Society Press, 1994.
White RD, Magnetic resonance imaging, In Topol EJ, editor. Textbook of Cardiovascular Medicine. Philadelphia, PA: Lippincott-Raven, 1998, pp. 1395-1445.
Sheehan FH, Bolson EL, Dodge HT, et al. Advantages and applications of the centerline method for characterizing regional ventricular function. Circulation 1986; 74(2): 293-305.
Staib LH, Duncan JS. Boundary finding with parametrically deformable models. IEEE Transac Pattern Analysis and Machine Intelligence 1992; 14: 1061-1075.
Horn BKP, Weldon EJ. Filtering closed curves. IEEE PAMI 1986; 8(5): 665-668.
Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 1986; i: 307-310.
Beyar R, Shapiro EP, Graves WL, et al. Quantification and validation of left ventricular wall thickening by a three-dimensionalvolume element magnetic resonance imaging approach. Circulation 1990; 81: 297-307.
Sechtem U, Sommerhoff BA, Markiewicz W, et al. Regional left ventricular wall thickening by magnetic resonance imaging: evaluation in normal persons and patients with global and regional dysfunction. Am J Cardiol 1987; 59: 145-151.
Ballard DH. Generalizing the Hough transform to detect arbitrary shapes. Pattern Recognition 1981; 13(2): 111-122.
Rueckert D, Burger P, Forbat SM, et al. Automatic tracking of the aorta in cardiovascular MR images using deformable models. IEEE Transac Med Imaging 1997; 15(5): 581-590.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Latson, L., Powell, K., Sturm, B. et al. Clinical validation of an automated boundary tracking algorithm on cardiac MR images. Int J Cardiovasc Imaging 17, 279–286 (2001). https://doi.org/10.1023/A:1011690219671
Issue Date:
DOI: https://doi.org/10.1023/A:1011690219671