Abstract
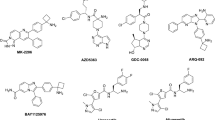
A three-dimensional quantitative structure-activity relationship (3D QSAR) study has been carried out on epothilones based on comparative molecular field analyses (CoMFA) using a large data set of epothilone analogs, which are potent inhibitors of tubulin depolymerization. Microtubules, which are polymers of the α/β-tubulin heterodimer, need to dissociate in order to form the mitotic spindle, a structure required for cell division. A rational pharmacophore searching method using 3D QSAR procedures was carried out and the results for the epothilones are described herein. One-hundred and sixty-six epothilone analogs and their depolymerization inhibition properties with tubulin were used as a training set. Over a thousand molecular field energies were generated and applied to generate the descriptors of QSAR equations. Using a genetic function algorithm (GFA) method, combined with a least square approach, multiple QSAR models were considered during the search for pharmacophore elements. Each GFA run resulted in 100 QSAR models, which were ranked according to their lack of fit (LOF) scores, with a total of 40 GFA runs having been performed. The 40 best QSAR equations from each run had adequate fitted correlation coefficients (R from 0.813 to 0.863) and were of sufficient statistical significance (F value from 7.2 to 10.9). The pharmacophore elements for epothilones were studied by investigating the hit frequency of descriptors (i.e. the sampling probabilities of grid points from the GFA studies) from the set of the 4000 top scoring QSAR equations. By comparing the frequency with which each grid point appeared in the QSAR equations, three candidate regions in the epothilones were proposed to be pharmacophore elements. Two of them are completely compatible with the recent model proposed by Ojima et al. [Proc. Natl. Acad. Sci. USA, 96 (1999) 4256], however, one is quite different and is necessary to accurately predict the activities of all 166 epothilone molecules used in our training set. Finally, by visualizing the 35 most probable grid points, it was found that changes related to the C6, C7, C8, C12, S20, and C21 atoms of the epothilones were highly correlated to their activity.
Similar content being viewed by others
References
Rwinsky, E.K., Cazene, L.C. and Donehower, R.C., J. Natl. Cancer Inst., 82 (1990) 1247.
Ojima, I., Kuduk, S.D. and Chakravarty, S., Adv. Med. Chem., 4 (1998) 69.
Ojima, I., Kuduk, S.D., Pera, P. and Veith, J.M., J. Med. Chem., 40 (1997) 279.
Shiff, P.B., Fant, J. and Horwitz, S.B., Nature, 277 (1979) 66.
Jordan, M.A., Toso, R.J. and Wilson, L., Proc. Natl. Acad. Sci. USA, 90 (1993) 9552.
Bollag, D.M., McQueney, P.A., Zhu, J., Hensens, O., Koupal, L., Liesch, J., Goetz, M., Lazarides, E. and Woods, C.M., Cancer Res., 55 (1995) 2325.
Kowalski, R.J., Giannakakou, P. and Hamel, E., J. Biol. Chem., 272 (1997) 2534.
Höfle, G., Bedorf, B., Gerth, K. and Reichenbach, H. (Gesellschaft für Biotechnologische Forschung, GBR), DE-B 4138042 1993 (Chem. Abstr., 120 (1993) 52841)
Gerth, K., Bedorf, N., Höfle, G. and Reichenbach, H., J. Antibiot., 49 (1996) 560.
Höfle, G., Bedorf, N., Steinmetz, H., Schomburg, D., Gerth, K. and Reichenbach, H., Angew. Chem. Int. Ed. Engl., 35 (1996) 1567.
Balog, A., Meng, D., Kamenecka, T., Bertinato, P., Su, D.-S., Sorensen, E.J. and Danishefsky, S.J., Angew. Chem. Int. Ed. Engl., 35 (1996) 2801.
Su, D.-S., Meng, D., Bertinato, P., Kamenecka, T., Balog, A., Sorensen, E.J., Danishefsky, S.J., Zheng, Y.-H., Chou, T.C., He, L. and Horwig, S.B., Angew. Chem. Int. Ed. Engl., 36 (1997) 757.
Yang, Z., He, Y., Vourloumis, D., Vallberg, H. and Nicolaou, K.C., Angew. Chem. Int. Ed. Engl., 36 (1997) 166.
Nicolaou, K.C., Sarabia, F., Ninkovic, S. and Yang, Z., Angew. Chem. Int. Ed. Engl., 36 (1997) 525.
Schinzer, D., Limberg, A., Bauer, A., Böhm, O.M. and Cordes, M., Angew. Chem. Int. Ed. Engl., 36 (1997) 523.
Martin, Y.C. and Willet, P. (Eds) Designing Bioactive Molecules: Three-Dimensional Techniques and Applications, American Chemical Society, Washington, DC, 1998.
Van De Waterbeemd, H., Advanced Computer-Assisted Techniques in Drug Discovery, VCH, Weinheim, New York, Basel, Cambridge, Tokyo, 1995.
Cramer III, R.D., Patterson, D.E. and Bunce, J.D., J. Am. Chem. Soc., 110 (1998) 5959.
Wold, S., Ruhe, A., Wold, H. and Dunn III, W.J., SIAM J. Sci. Stat. Comput., 5 (1984) 735.
Cramer, R.D., Bunce III, J.D. and Patterson, D.E., Quant. Struct.-Act. Relat., 7 (1988) 18.
Kulkarni, S.S. and Kulkarni, V.M., J. Med. Chem., 42 (1999) 373.
Holland, J. Adaptation in Artificial and Natural Systems, University of Michigan Press, Ann Arbor, MI, 1975.
Friedman, J., Multivariate Adaptive Regression Splines, Technical Report 102, Laboratory for Computational Statistics, Department of Statistics, Stanford University, Stanford, CA, 1988 (revised 1990)
Nicolaou, K.C., Vourloumis, D., Li, T., Pastor, J., Winssinger, N., He, Y., Ninkovic, S., Sarabia, F., Vallberg, H., Roschangar, F., King, N.P., Finlay, M.R.V., Giannakakou, P., Verdier-Pinard, P. and Hamel, E., Angew. Chem. Int. Ed. Engl., 36 (1997) 2093.
Nicolaou, K.C., Roschangar, F. and Vourloumis, D., Angew. Chem. Int. Ed. Engl., 37 (1998) 2014.
Halgren, T.A., J. Am. Chem. Soc., 114 (1992) 7827.
Halgren, T.A. and Nachbar, R.B., J. Compnt. Chem., 17 (1996) 587.
Halgren, T.A., J. Compnt. Chem., 20 (1999) 730.
Giannakakou, P., Gussio, R., Nogales, E., Downing, K.H., Zaharevitz, D., Bollbuck, B., Poy, G., Sackett, D., Nicolaou, K.C. and Fojo, T., Proc. Natl. Acad. Sci. USA, 97 (2000) 2904.
Sen, A. and Srivastava, M., Regression Analysis: Theory, Methods and Applications, Springer-Verlag, New York, NY, 1990.
Kachigan, S.K., Statistical Analysis: An Interdisciplinary Introduction to Univariate & Multivariate Methods, Radius Press, New York, NY, 1986.
Ojima, I., Chakravarty, S., Inoue, T., He, L., Horwitz, S.B., Kuduk, S.D. and Danishefsky, S.J., Proc. Natl. Acad. Sci. USA, 96 (1999) 4256.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Lee, K.W., Briggs, J.M. Comparative molecular field analysis (CoMFA) study of epothilones – tubulin depolymerization inhibitors: Pharmacophore development using 3D QSAR methods. J Comput Aided Mol Des 15, 41–55 (2001). https://doi.org/10.1023/A:1011140723828
Issue Date:
DOI: https://doi.org/10.1023/A:1011140723828