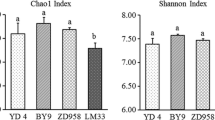
Abstract
The bacterial diversity and population dynamics in the rhizosphere of two maize cultivars (Nitroflint and Nitrodent) grown in tropical soils was studied, by traditional cultivation techniques and 16S rRNA gene-based molecular analysis of DNA directly extracted from soil and rhizosphere samples. Rhizosphere and soil samples were taken at three different plant growth stages. Total aerobic bacterial counts were determined. Fingerprints of the most dominant bacterial population were generated by TGGE separation of 16S rRNA gene fragments amplified from total community DNA using eubacterial specific primers. To reduce the complexity of TGGE fingerprints or to analyse less abundant populations, primers specific for different phylogenetic groups have been used. A comparison of the cfu obtained for rhizosphere of both cultivars indicated significant differences only for rhizosphere and soil samples taken 40 days after sowing. However, a comparison of TGGE patterns indicated that the composition of the bacterial community analysed at different plant growth stages for both cultivars was similar. A comparison of α-, β-proteobacterial and actinomycete TGGE patterns of both cultivars confirmed this observation. The eubacterial TGGE profiles reflected strong seasonal population shifts in the bacterial rhizosphere community of both maize cultivars which could be also observed in the TGGE patterns of α- and β-proteobacteria and to a lesser extent for actinomycetes. The rhizosphere effect was much more pronounced for young roots compared to samples taken from mature maize plants. The rhizosphere fingerprints showed a reduced complexity for young plants with up to five dominating bands while for mature plants patterns similar to those of soil were observed. Sequencing of dominant clones indicated that the dominant population found at all plant growth stages can be assigned to Arthrobacter populations.
Similar content being viewed by others
References
Amann R I, Ludwig W and Schleifer K-H 1995 Phylogenetic identi-fication and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59, 143–169.
Brim H, Heuer H, Krögerrecklenfort E, Mergeay M and Smalla K 1999 Characterization of the bacterial community of a zincpolluted soil. Can. J. Microbiol. 45, 326–338.
Chelius M K and Triplett E W 2000 Immunolocalization of dinitrogenase reductase produced by Klebsiella pneumoniae in association with Zea mays L. Appl. Environ. Microbiol. 66, 783–787.
Dalmastri C, Chiarini L, Cantale C, Bevivino A and Tabaccioni 1999 Soil type and maize cultivar affect the genetic diversity of maize root-associated Burkholderia cepacia populations. Microb. Ecol. 38, 273–284.
De Leij F A A M, Whipps J M and Lynch J M 1994 The use of colony development for the characterization of bacterial communities in soil and on roots. Microb. Ecol. 27, 81–97.
Di Cello F, Bevivino A, Chiarini L, Fani R, Paffetti D, Tabacchioni S and Dalmastri C 1997 Biodiversity of a Burkholderia cepacia population isolated from the maize rhizosphere at different plant growth stages. Appl. Environ. Microbiol. 63, 4485–4493.
Duineveld B M, Rosado A S, Van Elsas J D and Van Veen J A 1998 Analysis of the dynamics of bacterial communities in the rhizosphere of the Chrysanthemum via denaturing gradient gel electrophoresis and substrate utilization patterns. Appl. Environ. Microbiol. 64, 4950–4957.
Engelhard M, Hurek T and Reinhold-Hurek B 2000 Preferential occurrence of diazotrophic endophytes, Azoarcus spp., in wild rice species and land races of Oryza sativa in comparison with modern races. Environ. Microbiol. 2, 131–141.
Fogel G B, Collins C R, Li J and Brunk C F 1999 Prokaryotic genome size and SSU rDNA copy number. Microb. Ecol. 38, 93–113.
Gelsomino A, Keijzer-Wolters A, Cacco G and van Elsas J D 1999 Assessment of bacterial community structure in soil by polymerase chain reaction and denaturing gradient gel electrophoresis. J. Microbiol. Methods 38, 1–15.
Grayston S J, Wang S, Campbell C D and Edwards A C 1998 Selective influence of plant species on microbial diversity in the rhizosphere. Soil Biol. Biochem. 30, 369–378.
Heuer H and Smalla K 1997 Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) for studying soil microbial communities. In Modern soil microbiology. Eds J D van Elsas, E M H Wellington and J T Trevors. pp 353–373. Marcel Dekker, New York.
Heuer H, Krsek M, Baker P, Smalla K and Wellington E M H 1997 Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 63, 3233–3241.
Heuer H, Hartung K, Wieland G, Kramer I and Smalla K 1999 Polynucleotide probes that target a hypervariable region of 16S rRNA genes to identify bacterial isolates corresponding to bands of community fingerprints. Appl. Environ. Microbiol. 65, 1045–1049.
Höflich G, Wiehe W and Kühn G 1994 Plant growth stimulation by inoculation with symbiotic and associative rhizosphere microorganisms. Experientia 50 (Birkhäuser Verlag, Basel) 897–905.
Horwath W R, Elliott L F and Lynch J M 1998 Influence of soil quality on the function of inhibitory rhizobacteria. Lett. Appl. Microbiol. 26, 87–92.
Jaeger C H III, Lindow S E, Miller W, Clark E and Firestone M K 1999 Mapping of sugar and amino acid availability in soil around roots with bacterial sensors of sucrose and tryptophan. Appl. Environ. Microbiol. 65, 2685–2690.
Jones D and Keddie R M 1992 The Genus Arthrobacter. In The Prokaryotes, Vol. 2, 2nd edition. Eds A Balows, H G Trüpper, M Dworekin, W Harder and K-H Schleifer. pp 1283–1299, Springer, New York.
Klappenbach J A, Dunbar J M and Schmidt T M 2000 rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 66, 1328–1333.
Latour X, Philippot L, Corberand T and Lemanceau P 1996 The establishment of an introduced community of fluorescent pseudomonads in the soil and in the rhizosphere is affected by the soil type. FEMS Microbiol. Ecol. 30, 163–170.
Lilley, A K, Fry, J C, Bailey, M J and Day M J 1996 Comparison of aerobic heterotrophic taxa isolated from four root domains of mature sugar beet (Beta vulgaris). FEMS Microbiol. Ecol. 21, 231–242.
Lottmann, J, Heuer H, de Vries J, Mahn A, Düring K, Wackernagel W, Smalla K and Berg G 2000 Establishment of introduced antagonistic bacteria in the rhizosphere of transgenic potatoes and their effect on the bacterial community. FEMS Microb. Ecol. 33, 41–49.
Lupwayi N Z, Rice W A and Clayton GW 1998 Soil microbial diversity and community structure under wheat as influenced by tillage and crop rotation. Soil Biol. Biochem. 30, 1733–1741.
Mahaffee W F and Kloepper J W 1997 Temporal changes in the bacterial communities of soil, rhizosphere, and endorhiza associated with field grown cucumber (Cucumis sativus L.). Microb. Ecol. 34, 210–223.
Miller H J, Henken G and Van Veen J A 1989 Variation and composition of bacterial populations in the rhizospheres of maize, wheat, and grass cultivars. Can. J. Microbiol. 35, 656–660.
Muyzer G and Smalla K 1998 Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) inmicrobial ecology. Antonie van Leeuwenhoek 73, 127–141.
Muyzer G, de Waal E C and Uitterlinden A G 1993 Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes encoding for 16S rRNA. Appl. Environ. Microbiol. 59, 695–700.
Nübel U, Engelen B, Felske A, Snaidr J, Wiesenhuber A, Amann R I, Ludwig W and Backhaus H 1996 Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J. Bacteriol. 178, 5636–5643.
Picard C, Di Cello F, Ventura M, Fani R and Guckert A 2000 Frequency and biodiversity of 2,4-diacetylphloroglucinol-producing bacteria isolated from the maize rhizosphere at different stages of plant growth. Appl. Environ. Microbiol. 66, 948–955.
Riesner D, Steger G, Zimmat R, Owens R A, Wagenhöfer M, Hillen W, Vollbach S and Henco K 1989 Temperature-gradient gel electrophoresis of nucleic acids: analysis of conformational transitions, sequence variations, and protein–nucleic acid interactions. Electrophoresis 10, 377–389.
Rychlik W and Rhoads R E 1989 A computer program for choosing optimal oligonucleotides for filter hybridization, sequencing and in vitro amplification of DNA. Nucleic Acids Res. 17, 8543–8551.
Sambrook J, Fritsch E F and Maniatis T 1989 Molecular Cloning: a Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
Seldin L, Rosado A S, Da Cruz D W, Nobrega A, Van Elsas J D and Paiva E 2000 Comparison of Paenibacillus azotofixans strains isolated from rhizoplane, rhizosphere, and non-root-associated soil from maize planted in two different Brazilian soils. Appl. Environ. Microbiol. 64, 3860–3868.
Sheffield, V C, Cox R D, Lerman L S and Myers R M 1989. Attachment of a 40-base-pair G+C-rich sequence (GC-clamp) to genomic DNA fragments by the polymerase chain reaction results in improved detection of single-base changes. Proc. Natl. Acad. Sci. USA 86, 232–236.
Smalla K and Van Elsas J D 1995 Application of the PCR for detection of antibiotic resistance genes in environmental samples. In Nucleic Acids in the Environment. Eds J T Trevors, J D van Elsas. pp 241–256. Springer, Berlin, Heidelberg.
Smalla K, Cresswell N, Mendonca-Hagler L C, Wolters A and Van Elsas J D 1993 Rapid DNA extraction protocol from soil for polymerase chain reaction-mediated amplification. J. Appl. Bacteriol. 74, 78–85.
Staley, J T and Konopka A 1985. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu. Rev. Microbiol. 39, 321–346.
Suzuki M, Rappé M S and Giovannoni S J 1998 Kinetic bias in estimates of coastal picoplankton community structure obtained by measurements of small-subunit rRNA gene PCR amplicon length heterogeneity. Appl. Environ. Microbiol. 64, 4522–4529.
Van Elsas, J D and Smalla K 1996 Methods for sampling soil microbes. In Manual of Environmental Microbiology. Eds C J Hurst, G R Knudsen, M J McInerney, L D Stetzenbach and M V Walter. pp 383–390. ASM Press, Washington, DC.
Van Loon L C, Bakker P A H M and Pieterse C M J 1998 Systemic resistance induced by rhizosphere bacteria. Annu. Rev. Phytopathol. 36, 453–483.
von Wintzingerode F, Göbel U B and Stackebrandt E 1997 Determination of Microbial Diversity in Environmental Samples: Pitfalls of PCR-based rRNA Analysis. FEMS Microbiol. Rev. 21, 213–229.
Watkinson A R 1998 The role of the soil community in plant population dynamics. TREE 13, 171–172.
Weisburg W G, Barns S M, Pelletier D A and Lane D J 1991 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703.
Westover K M, Kennedy A C and Kelley S E 1997 Patterns of rhizosphere microbial community structure associated with co-occurring plant species. J. Ecol. 85, 863–873.
Wilson K 1987 Preparation of genomic DNA from bacteria. In Current Protocols in Molecular Biology. Eds J A Ausubel, R Bent, R E Kingston, D D Moore, J A Smith, J G Seidmann and K Struhl. pp 2.10–2.12. Wiley, New York, NY.
Yang C-H and Crowley D E 2000 Rhizosphere microbial community structure in relation to root location and plant iron nutritional status. Appl. Environ. Microbiol. 66, 345–351.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gomes, N.C.M., Heuer, H., Schönfeld, J. et al. Bacterial diversity of the rhizosphere of maize (Zea mays) grown in tropical soil studied by temperature gradient gel electrophoresis. Plant and Soil 232, 167–180 (2001). https://doi.org/10.1023/A:1010350406708
Issue Date:
DOI: https://doi.org/10.1023/A:1010350406708