Abstract
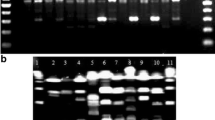
Typing of reference strains and isolates identified as Lactobacillus casei, Lactobacillus paracasei or Lactobacillus rhamnosus was carried out using randomly amplified polymorphic DNA (RAPD) and pulsed-field gel electrophoresis (PFGE) analyses. Strains of L. paracasei were mainly grouped in the same cluster as those of L. casei. The RAPD fingerprints of strains ATCC 393 and ATCC 15820 differ from those of the L. rhamnosus and L. paracasei/casei strains further supporting classification of these strains as a separate group. The RAPD profiling could be used for classification and discrimination of isolates belonging to the L. casei group.
Similar content being viewed by others
References
Collins MD, Phillips BA, Zanoni P (1989) Deoxyribonucleic acid homology studies of Lactobacillus casei, Lactobacillus paracasei sp. nov., subsp. paracasei and subsp. tolerans, and Lactobacillus rhamnosus sp. nov., comb. nov. Int. J. Syst. Bacteriol. 39: 105-108.
Daud Khaled AK, Neilan BA, Henriksson A, Conway PL (1997) Identification and phylogenetic analysis of Lactobacillus using multiplex RAPD-PCR. FEMS Microbiol. Lett. 153: 191-197.
Dellaglio F, Bottazzi V, Vescovo M (1975) Deoxyribonucleic acid homology among Lactobacillus species of the subgenusStreptobacterium Orla-Jensen. Int. J. Syst. Bacteriol. 25: 160-172.
Dellaglio F, Dicks LMT, Du Toit M, Torriani S (1991) Designation of ATCC 334 in place of ATCC 393 (NCDO 161) as the neotype strain of Lactobacillus casei subsp. casei and rejection of the name Lactobacillus paracasei (Collins et al. 1989). Int. J. Syst. Bacteriol. 25: 160-172.
Dicks LMT, Du Plessis EM, Dellaglio F, Lauer E (1996) Reclassification of Lactobacillus rhamnosus ATCC 15820 as Lactobacillus zeae nom. rev., designation of ATCC 334 as neotype of L. casei subsp. casei, and rejection of the name Lactobacillus paracasei. Int. J. Syst. Bacteriol. 46: 337-340.
Du Plessis EM, Dicks LMT (1995) Evaluation of random amplified polymorphic DNA (RAPD)-PCR as a method to differentiate Lactobacillus acidophilus, Lactobacillus crispatus, Lactobacillus amylovorus, Lactobacillus gallinarum, Lactobacillus gasseri, and Lactobacillus johnsonii. Curr. Microbiol. 31: 114-118.
Dykes GA, von Holy A (1994) Strain typing in the genus Lactobacillus. Lett. Appl. Microbiol. 19: 63-66.
Johannson ML, Quednau M, Molin G, Ahrné S (1995) Randomly amplified polymorphic DNA (RAPD) for rapid typing of Lactobacillus plantarum strains. Lett. Appl. Microbiol. 21: 155-159.
Klein G, Pack A, Bonaparte C, Reuter G (1998) Taxonomy and physiology of probiotic lactic acid bacteria. Int. J. Food Microbiol. 41: 103-125.
Lortal S, Rouault A, Guezenec S, Gautier M (1997) Lactobacillus helveticus: strain typing and genome size estimation by pulsed field gel electrophoresis. Curr. Microbiol. 34: 180-185.
Mori K, Yamazaki K, Ishiyama T, Katsumata M, Kobayashi K, Kawai Y, Inoue N, Shinano H (1997) Comparative sequence analyses of the genes coding for 16S rRNA of Lactobacillus casei-related taxa. Int. J. Syst. Bacteriol. 47: 54-57.
Roy D, Ward P, Champagne G (1996) Differentiation of bifidobacteria by use of pulsed-field gel electrophoresis and polymerase chain reaction. Int. J. Food Microbiol. 29: 11-29.
Salminen S, Isolauri E, Salminen E (1996) Clinical uses of probiotics for stabilizing the gut mucosal barrier: successful strains and future challenges. Antonie van Leeuwenhoek 70: 347-358.
Sambrook J, Fristch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, 2nd ed. Cold Spring Harbor, MA: Cold Spring Harbor Laboratory Press.
Taillez P, Quénée P, Chopin A (1996) Estimation de la diversité parmi les souches de la collection CNRZ: application de la RAPD à un groupe de lactobacilles. Lait 76: 147-158.
Tilsala-Timisjärvi A, Alatossava T (1998) Strain-specific identification of probiotic Lactobacillus rhamnosus with randomly amplified polymorphic DNA-derived PCR primers. Appl. Environ. Microbiol. 64: 4816-4819.
Vincent D, Roy D, Mondou F, Déry C (1998) Characterization of bifidobacteria by random DNA amplification. Int. J. Food Microbiol. 43: 185-193.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Roy, D., Ward, P. & Vincent, D. Strain identification of probiotic Lactobacillus casei-related isolates with randomly amplified polymorphic DNA and pulsed-field gel electrophoresis methods. Biotechnology Techniques 13, 843–847 (1999). https://doi.org/10.1023/A:1008952023716
Issue Date:
DOI: https://doi.org/10.1023/A:1008952023716