Abstract
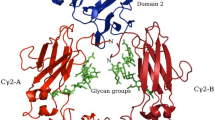
A molecular model of the complex between Fas and its ligand was generated to better understand the location and putative effects of site-specific mutations, analyze interactions at the Fas–FasL interface, and identify contact residues. The modeling study was conservative in the sense that regions in Fas and its ligand which could not be predicted with confidence were omitted from the model to ensure accuracy of the analysis. Using the model, it was possible to map four of five N-linked glycosylation sites in Fas and FasL and to study 10 of 11 residues previously identified by mutagenesis as important for binding. Interactions involving six of these residues could be analyzed in detail and their importance for binding was rationalized based on the model. The predicted structure of the Fas–FasL interface was consistent with the experimentally established importance of these residues for binding. In addition, five previously not targeted residues were identified and predicted to contribute to binding via electrostatic interactions. Despite its limitations, the study provided a much improved basis to understand the role of Fas and FasL residues for binding compared to previous residue mapping studies using only a molecular model of Fas.
Similar content being viewed by others
References
Itoh, N., Yonehara, S., Ishii, A., Yonehara, M., Mizushima, S.-I., Sameshima, M., Hase, A., Seto, Y. and Nagata, S., Cell, 66 (1991) 233.
Suda, T., Takahashi, T., Golstein, P. and Nagata, S., Cell, 75 (1994) 1169.
Beutler, B. and van Huffel, C., Science, 264 (1994) 667.
Smith, C.A., Farrah, T. and Goodwin, R.G., Cell, 76 (1994) 959.
Nagata, S. and Golstein, P., Science, 267 (1995) 1449.
Nagata, S., Adv. Exp. Med. Biol., 406 (1996) 119.
Eck, M.J. and Sprang, S.R., J. Biol. Chem., 264 (1989) 17595.
Eck, M.J., Ultsch, M., Rinderknecht, E., de Vos, A.M. and Sprang, S.R., J. Biol. Chem., 267 (1992) 2119.
Karpusas, M., Hsu, Y.-M., Wang, J.-H., Thompson, J., Lederman, S., Chess, L. and Thomas, D., Structure, 3 (1995) 1031.
Naismith, J.H., Devine, T.Q., Brandhuber, B.J. and Sprang, S.R., J. Biol. Chem., 270 (1995) 13303.
Banner, D.W., D'Arcy, A., Janes, W., Gentz, R., Schoenfeld, H.-J., Broger, C., Loetscher, H. and Lesslauer, W., Cell, 73 (1993) 431.
Naismith, J.H. and Sprang, S.R., Trends Biochem. Sci., 23 (1998) 74.
Bajorath, J., Stenkamp, R. and Aruffo, A., Protein Sci., 2 (1993) 1798.
Bajorath, J. and Aruffo, A., Proteins Struct. Funct. Genet., 27 (1997) 59.
Bajorath, J. and Aruffo, A., J. Comput.-Aided Mol. Design, 11 (1997) 3.
Bajorath, J., J. Mol. Model., 4 (1998) 239. 17.
Peitsch, M.C. and Jongeneel, C.V., Int. Immunol., 5 (1993) 233.
Bajorath, J., Marken, J.S., Chalupny, N.J., Spoon, T.L., Siadak, A.W., Gordon, M., Noelle, R.J., Hollenbaugh, D. and Aruffo, A., Biochemistry, 34 (1995) 9884.
Peitsch, M.C. and Tschopp, J., J. Mol. Immunol., 32 (1995) 761.
Singh, J., Garber, E., Van Vlijmen, H., Karpusas, M., Hsu, Y.-M., Zheng, Z., Naismith, J.H. and Thomas, D., Protein Sci., 7 (1998) 1124.
Schneider, P., Bodmer, J.-L., Holler, N., Mattmann, C., Scuderi, P., Terskikh, A., Peitsch, M.C. and Tschopp, J., J. Biol. Chem., 272 (1998) 18827.
van Ostade, X., Tavernier, J. and Fiers, W., Protein Eng., 7 (1994) 5.
Starling, G.C., Bajorath, J., Emswiler, J., Ledbetter, J.A., Aruffo, A. and Kiener, P.A., J. Exp. Med., 185 (1997) 1487.
Starling, G.C., Kiener, P.A., Aruffo, A. and Bajorath, J., Biochemistry, 37 (1998) 3723.
Martin, A.C., MacArthur, M.W. and Thornton, J.M., Proteins Struct. Funct. Genet. Suppl., 1 (1998) 14.
Bajorath, J., J. Biol. Chem., 273 (1998) 24603.
Needleman, S.B. and Wunsch, C.D., J. Mol. Biol., 48 (1970) 443.
Gonnet, G.H., Cohen, M.A. and Benner, S.A., Science, 256 (1992) 1433.
Bairoch, A. and Apwiler, R., Nucleic Acids Res., 26 (1998) 38.
Bernstein, F.C., Koetzle, T.F., Williams, G.J.B., Meyer, E.F., Jr., Brice, M.D., Rodgers, J.R., Kennard, O., Shimanouchi, T. and Tasumi, M., J. Mol. Biol., 112 (1977) 535.
Ponder, J. W. and Richards, F. M., J. Mol. Biol., 193 (1987) 775.
Bajorath, J. and Fine, R. M., Immunomethods, 1 (1992) 137.
Laskowski, R.A., MacArthur, M.W., Moss, D.S. and Thornton, J.M., J. Appl. Crystallogr., 26 (1993) 283.
Bruccoleri, R.E. and Novotny, J., Immunomethods, 1 (1992) 96.
Peitsch, M. C., Biotechnology, 13 (1995) 658.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Bajorath, J. Analysis of Fas–ligand interactions using a molecular model of the receptor–ligand interface. J Comput Aided Mol Des 13, 409–418 (1999). https://doi.org/10.1023/A:1008031200961
Issue Date:
DOI: https://doi.org/10.1023/A:1008031200961