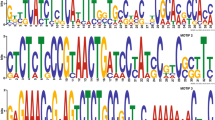
Abstract
We have isolated cDNA clones encoding class I chitinase (ChtC) from potato leaves which share a high degree of nucleotide and amino acid sequence similarity to other, previously described basic (class I) chitinases (ChtB) from potato. Despite this similarity, characteristic features distinguish ChtC from ChtB, including an extended proline-rich linker region between the hevein and catalytic domains and presence of a potential glycosylation site (NDT) in the deduced protein. These differences are in accordance with the properties of purified chitinase C which is glycosylated and hence has a higher molecular mass in comparison to chitinase B. In contrast to the coding sequences, the 3′-untranslated regions of ChtC and ChtB exhibited a low degree of similarity, which allowed us to generate gene-specific probes to study the genomic organization and expression of both types of gene. Genomic DNA blots suggest that ChtC and ChtB are each encoded by one or two genes per haploid genome. RNA blot analysis showed that in healthy potato plants ChtC mRNA is most abundant in young leaves, the organs which also contain high levels of chitinase C. By contrast, ChtB mRNA abundance is highest in old leaves, which accumulate chitinase B. By in situ RNA hybridization with gene-specific probes we could demonstrate that ChtC mRNA in leaves is restricted to epidermal cells, whereas ChtB mRNA showed no distinct pattern of cell-type-specific localization. Infection of potato leaves with Phytophthora infestans, or treatment with fungal elicitor, ethylene, or wounding resulted in accumulation of both ChtC and ChtB mRNAs; however, for ChtC, in contrast to ChtB, no corresponding accumulation of the encoded protein could be detected, suggesting a post-transcriptional mechanism of regulation. Salicylic acid treatment did not induce accumulation of either mRNA. The possible functional implications of these findings for pathogen defence and developmental processes are discussed.
Similar content being viewed by others
References
Abeles FB, Bosshart RP, Forrence LE, Habig WH: Preparation and purification of glucanase and chitinase from bean leaves. Plant Physiol 47: 129–134 (1971).
Bartnicki-Garcia S: Cell wall chemistry, morphogenesis, and taxonomy of fungi. Annu Rev Microbiol 22: 87–108 (1968).
Beerhues L, Kombrink E: Primary structure and expression of mRNAs encoding basic chitinase and 1,3-β-glucanase in potato. Plant Mol Biol 24: 353–367 (1994).
Boller T: Ethylene and the regulation of antifungal hydrolases in plants. In: Miflin BJ (ed) Oxford Surveys of PlantMolecular and Cell Biology, vol 5, pp. 145–174. Oxford University Press, Oxford (1988).
Brederode FT, Linthorst HJM, Bol JF: Differential induction of acquired resistance and PR gene expression in tobacco by virus infection, ethephon treatment, UV light and wounding. Plant Mol Biol 17: 1117–1125 (1991).
Broekaert W, Lee H-I, Kush A, Chua N-H, Raikhel N:Woundinduced accumulation of mRNA containing a hevein sequence in laticifers of rubber tree (Hevea brasiliensis). Proc Natl Acad Sci USA 87: 7633–7637 (1990).
Broglie K, Biddle P, Cressman R, Broglie R: Functional analysis of DNA sequences responsible for ethylene regulation of a bean chitinase gene in transgenic tobacco. Plant Cell 1: 599–607 (1989).
Büchter R, Strömberg A, Schmelzer E, Kombrink E: Primary structure and expression of acidic (class II) chitinase in potato. Plant Mol Biol 35: 749–761 (1997).
Chen EY, Seeburg PH: Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA 4: 165–170 (1985).
Collinge DB, Kragh KM, Mikkelsen JD, Nielsen KK, Rasmussen U, Vad K: Plant chitinases. Plant J 3: 31–40 (1993).
Dellaporta SL, Wood J, Hicks JB: A plant DNA minipreparation: version II. Plant Mol Biol Rep 1: 19–21 (1983).
Devereux J, Haeberli P, Smithies O: A comprehensive set of sequence analysis programs for the VAX. Nucl Acids Res 12: 387–395 (1984).
Eyal Y, Fluhr R: Cellular and molecular biology of pathogenesis-related proteins. Oxford Surv Plant Mol Cell Biol 7: 223–254 (1991).
Frohmann MA, Dush MK, Martin GR: Rapid amplification of full-length cDNAs from rare transcripts: Amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA 85: 8998–9002 (1988).
Garcia-Garcia F, Schmelzer E, Hahlbrock K, Roxby R: Differential expression of chitinase and β-1,3-glucanase genes in various tissues of potato plants. Z Naturforsch 49c: 195–203 (1994).
Graham LS, Sticklen MB: Plant chitinases. Can J Bot 72: 1057–1083 (1994).
Jach G, Görnhardt B, Mundy J, Logemann J, Pinsdorf E, Leah R, Schell J, Maas C: Enhanced quantitative resistance against fungal disease by combinatorial expression of different barley antifungal proteins in transgenic tobacco. Plant J 8: 97–109 (1995).
Joshi CP: Putative polyadenylation signals in nuclear genes of higher plants: a compilation and analysis. Nucl Acids Res 15: 9627–9640 (1987).
Keefe D, Hinz U, Meins F Jr: The effect of ethylene on the cell-type-specific and intracellular localization of β-1,3-glucanase and chitinase in tobacco leaves. Planta 182: 43–51 (1990).
Kombrink E, Beerhues L, Garcia-Garcia F, Hahlbrock K, Müller M, Schröder M, Witte B, Schmelzer E: Expression patterns of defense-related genes in infected and uninfected plants. In: Fritig B, Legrand M (eds) Mechanisms of Plant Defense Responses, vol 2, pp. 236–249. Kluwer Academic Publishers, Dordrecht, Netherlands (1993).
Kombrink E, Schröder M, Hahlbrock K: Several 'pathogenesis-related' proteins in potato are 1,3-β-glucanases and chitinases. Proc Natl Acad Sci USA 85: 782–786 (1988).
Kombrink E, Somssich IE: Defense responses of plants to pathogens. In: Andrews JH, Tommerup IC (eds) Advances in Botanical Research (incorporating Advances in Plant Pathology), vol 21, pp. 1–34. Academic Press, London (1995).
Kombrink E, Somssich IE: Pathogenesis-related proteins and plant defense. In: Carroll G, Tudzynski P (eds) The Mycota, Vol 5 Part A, Plant Relationships, pp. 107–128. Springer-Verlag, Berlin/Heidelberg (1997).
Lamb CJ, Ryals JA, Ward ER, Dixon RA: Emerging strategies for enhancing crop resistance to microbial pathogens. Bio/technology 10: 1436–1445 (1992).
Lauriè re M, Lauriè re C, Chrispeels MJ, Johnson KD, Sturm A: Characterization of a xylose-specific antiserum that reacts with the complex asparagine-linked glycans of extracellular and vacuolar glycoproteins. Plant Physiol 90: 1182–1188 (1989).
Leah R, Tommerup H, Svendsen I, Mundy J: Biochemical and molecular characterization of three barley seed proteins with antifungal properties. J Biol Chem 266: 1564–1573 (1991).
Linthorst HJM, van Loon LC, van Rossum CMA, Mayer A, Bol JF, van Roekel JSC, Meulenhoff EJS, Cornelissen BJC: Analysis of acidic and basic chitinases from tobacco and petunia an their constitutive expression in transgenic tobacco. Mol Plant-Microbe Interact 3: 252–258 (1990).
Lotan T, Ori N, Fluhr R: Pathogenesis-related proteins are developmentally regulated in tobacco flowers. Plant Cell 1: 881–887 (1989).
Mauch F, Mauch-Mani B, Boller T: Antifungal hydrolases in pea tissue. II. Inhibition of fungal growth by combinations of chitinase and 1,3-β-glucanase. Plant Physiol 88: 936–942 (1988).
Mauch F, Meehl JB, Staehelin LA: Ethylene-induced chitinase and β-1,3-glucanase accumulate specifically in the lower epidermis and along vascular strands of bean leaves. Planta 186: 367–375 (1992).
Meins F Jr, Neuhaus J-M, Sperisen C, Ryals J: The primary structure of plant pathogenesis-related glucanohydrolases and their genes. In: Boller T, Meins F Jr (eds) Genes Involved in Plant Defense, pp. 245–282. Springer-Verlag, Vienna (1992).
Melchers LS, Apotheker-de Groot M, van der Knaap JA, Ponstein AS, Sela-Buurlage MB, Bol JF, Cornelissen BJC, van den Elzen PJM, Linthorst HJM: A new class of tobacco chitinase homologous to bacterial exo-chitinases displays antifungal activity. Plant J 5: 469–480 (1994).
Memelink J, Linthorst HJM, Schilperoort RA, Hoge JHC: Tobacco genes encoding acidic and basic isoforms of pathogenesis-related proteins display different expression patterns. Plant Mol Biol 14: 119–126 (1990).
Neuhaus J-M, Sticher L, Meins F Jr, Boller T: A short Cterminal sequence is necessary and sufficient for the targeting of chitinases to the plant vacuole. Proc Natl Acad Sci USA 88: 10362–10366 (1991).
Ori N, Sessa G, Lotan T, Himmelhoch S, Fluhr R: Amajor stylar matrix polypeptide (sp41) is a member of the pathogenesisrelated proteins superclass. EMBO J 9: 3429–3436 (1990).
Samac DA, Hironaka CM, Yallaly PE, Shah DM: Isolation and characterization of the genes encoding basic and acidic chitinase in Arabidopsis thaliana. Plant Physiol 93: 907–914 (1990).
Samac DA, Shah DM: Developmental and pathogen-induced activation of the Arabidopsis acidic chitinase promoter. Plant Cell 3: 1063–1072 (1991).
Sambrook J, Fritsch EF, Maniatis T: Molecular Cloning: A Laboratory Manual, 2nd ed. Cold Spring Harbor Laboratory Press, Plainview, NY (1989).
Schröder M, Hahlbrock K, Kombrink E: Temporal and spatial patterns of 1,3-β-glucanase and chitinase induction in potato leaves infected by Phytophthora infestans. Plant J 2: 161–172 (1992).
Sela-Buurlage MB, Ponstein AS, Bres-Vloemans SA, Melchers LS, van den Elzen PJM, Cornelissen BJC: Only specific tobacco (Nicotiana tabacum) chitinases and β-1,3-glucanases exhibit antifungal activity. Plant Physiol 101: 857–863 (1993).
Shinshi H, Mohnen D, Meins F Jr: Regulation of a plant pathogenesis-related enzyme: inhibition of chitinase and chitinase mRNA accumulation in cultured tobacco tissues by auxin and cytokinin. Proc Natl Acad Sci USA 84: 89–93 (1987).
Somssich IE: Regulatory elements governing pathogenesisrelated (PR) gene expression. In: Nover L (ed), Results and Problems in Cell Differentiation, vol 20, pp. 163–179. Springer-Verlag, Berlin/Heidelberg (1994).
Sticher L, Hofsteenge J, Neuhaus J-M, Boller T, Meins F Jr: Posttranslational processing of a new class of hydroxyprolinecontaining proteins. Prolyl hydroxylation and C-terminal cleavage of tobacco (Nicotiana tabacum) vacuolar chitinase. Plant Physiol 101: 1239–1247 (1993).
van Hengel AJ, Guzzo F, van Kammen A, de Vries SC: Expression pattern of the carrot EP3 endochitinase genes in suspension cultures and in developing seeds. Plant Physiol 117: 43–53 (1998).
van Holst G-J, Varner JE: Reinforced polyprolin II conformation in a hydroxyproline-rich cell wall glycoprotein from carrot root. Plant Physiol 74: 247–251 (1984).
von Heijne G: Patterns of amino acids near signal-sequence cleavage sites. Eur J Biochem 133: 17–21 (1983).
Ward ER, Uknes SJ, Williams SC, Dincher SS, Wiederhold DL, Alexander DC, Ahl-Goy P, Metraux J-P, Ryals JA: Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell 3: 1085–1094 (1991).
Wemmer T, Kaufmann H, Kirch H-H, Schneider K, Lottspeich F, Thompson RD: The most abundant soluble basic protein of the stylar transmitting tract in potato (Solanum tuberosum L.) is an endochitinase. Planta 194: 264–273 (1994).
Wessels JGH, Sietsma JH: Fungal cell walls: a survey. In: Tanner W, Loewus FA (eds) Encyclopedia of Plant Physiology, New Series, vol 13B, Plant Carbohydrates II, pp. 352–394. Springer-Verlag, Berlin (1981).
Witte B, Hahlbrock K, Kombrink E: Purification, characterization of tree types of chitinase from potato leaves that differ in expression and cellular localization. In preparation.
Zhu Q, Doerner PW, Lamb CJ: Stress induction and developmental regulation of a rice chitinase promoter in transgenic tobacco. Plant J 3: 203–212 (1993).
Zhu Y, Maher EA, Masoud S, Dixon RA, Lamb CJ: Enhanced protection against fungal attack by constitutive coexpression of chitinase and glucanase genes in transgenic tobacco. Bio/technology 12: 807–812 (1994).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Ancillo, G., Witte, B., Schmelzer, E. et al. A distinct member of the basic (class I) chitinase gene family in potato is specifically expressed in epidermal cells. Plant Mol Biol 39, 1137–1151 (1999). https://doi.org/10.1023/A:1006178425803
Issue Date:
DOI: https://doi.org/10.1023/A:1006178425803