Abstract
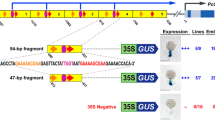
MNF1 is a factor which specifically binds to a 318 bp fragment (-1012 to -695) in the 5′-flanking region of the C4-type phosphoenolpyruvate carboxylase gene in Zea mays (Yanagisawa et al., Mol Gen Genet 224 (1990) 325–332). The most preferred binding site of MNF1 determined by a 2 bp mutation-scanning assay was an octamer sequence, GTGCCCTT, which is located within the repeated sequences (RS1; -886 to -849, -846 to -807). Furthermore, a PCR-mediated selection-amplification assay identified both the octamer sequence, GTGCCC(A/T)(A/T), and an additional sequence, CC(G/A)CCC, the latter of which was similar to the Sp1 sites in vertebrates. Specific binding of MNF1 to each of the supposed binding sites was confirmed with double-stranded monomers as probes. Considering native molecular mass of MNF1 (ca. 500 kDa), a protein complex is expected. In addition, MNF1 is anticipated to have two distinct DNA-binding proteins since the MNF1 binding to CCGCCC element was 1,10-phenanthroline-dependent whereas the MNF1 binding to the octamer was independent. Wide distribution of the MNF1 binding sequences within the 1 kb promoter region accounts for broad interactions of MNF1. Moreover, specific DNA binding due to MNF1, which was not observed in the nuclear extract derived from germinated and cultivated plants in darkness, appeared after a white-light pulse. This finding suggests the involvement of the protein complex in the light-dependent transcriptional control in the gene expression.
Similar content being viewed by others
References
Andreo CS, Gonzalez DH, Iglesias AA: Higher plant phosphoenolpyruvate carboxylase. FEBS Lett 213: 1–8 (1987).
Blackwell TK, Weintraub H: Differences and similarities in DNA binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science 250: 1104–1110 (1990).
Briggs MR, Kadonaga JT, Bell SP, Tjian R: Purification and biochemical characterization of the promoter-specific transcription factor, Sp1. Science 234: 47–52 (1986).
Bruce WB, Deng X-W, Quail PH: A negatively acting DNA sequence element mediates phytochrome-directed repression of phyA gene transcription. EMBO J 10: 3015–3024 (1991).
Chamovitz DA, Wei N, Osterlund MT, von Arnim AG, Staub JM, Matsui M, Deng X-W: The COP9 complex, a novel multisubunit nuclear regulator involved in light control of a plant developmental switch. Cell 86: 115–121 (1996).
Christensen AH, Quail PH: Structure and expression of a maize phytochrome-encoding gene. Gene 85: 381–390 (1989).
Fischer KD, Haese A, Nowock J: Cooperation of GATA-1 and Sp1 can result in synergistic transcriptional activation or interference. J Biol Chem 268: 23915–23923 (1993).
Green PJ, Kay SA, Lam E, Chua N-H: in vitro DNA footprinting. In: Gelvin SB, Schilperoort RA (eds) Plant Molecular Biology Manual, B11: 7–10. Kluwer Academic Publishers Dordrecht, Netherlands (1989).
Goodrich JA, Cutler G, Tjian R: Contacts in context: promoter specificity and macromolecular interactions in transcription. Cell 84: 825–830 (1996).
Hayakawa S, Matsunaga K, Sugiyama T: Light induction of phosphoenolpyruvate carboxylase in etiolated maize leaf tissue. Plant Physiol 67: 133–138 (1981).
Hiratsuka K, Wu X, Fukuzawa H, Chua N-H: Molecular dissection of GT-1 from Arabidopsis. Plant Cell 6: 1805–1813 (1994).
Kadonaga JT, Carner KR, Masiarz FR, Tjian R: Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell 51: 1079–1090 (1987).
Kano-Murakami Y, Suzuki I, Sugiyama T, Matsuoka M: Sequence-specific interactions of a maize factor with a GCrich repeat in the phosphoenolpyruvate carboxylase gene. Mol Gen Genet 225: 203–208 (1991).
Kuhn RM, Caspar T, Dehesh K, Quail PH: DNAbinding factor GT-2 from Arabidopsis. Plant Mol Biol 23: 337–348 (1993).
Langdale JA, Nelson T: Spatial regulation of photosynthetic development in C4 plants. Trends Genet 7: 191–196 (1991).
Lee J-S, Galvin KM, Shi Y: Evidence for physical interaction between the zinc-finger transcription factors YY1 and Sp1. Proc Natl Acad Sci USA 90: 6145–6149 (1993).
Maniatis T, Fritsch EF, Sambrook J: Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY (1982).
Mastrangelo IA, Courey AJ, Wall JS, Jackson SP, Hough PVC: DNA looping and Sp1 multimer links: a mechanism for transcriptional synergism and enhancement. Proc Natl Acad Sci USA 88: 5670–5674 (1991).
Matsuoka M, Minami E: Complete structure of the gene for phosphoenolpyruvate carboxylase from maize. Eur J Biochem 181: 593–598 (1989).
Merika M, Orkin SH: Functional synergy and physical interactions of the erythroid transcription factor GATA-1 with the Krüppel family proteins Sp1 and EKLF. Mol Cell Biol 15: 2437–2447 (1995).
Nelson T, Harpster MH, Mayfield SP, Taylor WC: Lightregulated gene expression during maize leaf development. J Cell Biol 98: 558–564 (1984).
Pascal E, Tjian R: Different activation domains of Sp1 govern formation ofmultimers and mediate transcriptional synergism. Genes Devel 5: 1646–1656 (1991).
Perkins ND, Edwards NL, Duckett CS, Agranoff AB, Schmid RM, Nabel GJ: A cooperative interaction between NF-kB and Sp1 is required for HIV-1 enhancer activation. EMBO J 12: 3551–3558 (1993).
Perrot-Rechenmann C, Vidal J, Brulfert J, Burlet A, Gadal P: A comparative immunocytochemical localization study of phosphoenolpyruvate carboxylase in leaves of higher plants. Planta 155: 24–30 (1982).
Roeder RG, Rutter WJ: Specific nucleolar and nucleoplasmic RNA polymerases. Proc Natl Acad Sci USA 65: 675–682 (1970).
Schäffner AR, Sheen J: Maize C4 photosynthesis involves differential regulation of phosphoenolpyruvate carboxylase gene. Plant J 2: 221–232 (1992).
Sheen J-Y, Bogorad L: Differential expression of C4 pathway genes in mesophyll and bundle sheath cells of greening maize leaves. J Biol Chem 262: 11726–11730 (1987).
Sims TL, Hague DR: Light-stimulated increase of translatable mRNA for phosphoenolpyruvate carboxylase in leaves of maize. J Biol Chem 256: 8252–8255 (1981).
Sun J-M, Penner CG, Davie JR: Analysis of erythroid nuclear proteins binding to the promoter and enhancer elements of the chicken histone H5 gene. Nucl Acids Res 20: 6385–6392 (1992).
Thiesen H-J, Bach C: Target detection assay (TDA): a versatile procedure to determine DNA binding sites as demonstrated on SP1 protein. Nucl Acids Res 18: 3203–3209 (1990).
Thomas M, Cretin C, Vidal J, Keryer E, Gadal P, Monsinger E: Light regulation of phosphoenolpyruvate carboxylase mRNA in leaves of C4 plants: evidence for phytochrome control on transcription during greening and for rhythmicity. Plant Sci 69: 65–78 (1990).
Yanagisawa S, Izui K: Maize phosphoenolpyruvate carboxylase involved in C4 photosynthesis: nucleotide sequence analysis of the 50 flanking region of the gene. J Biochem 106: 982–987 (1989).
Yanagisawa S, Izui K: Multiple interactions between tissuespecific nuclear proteins and the promoter of the phosphoenolpyruvate carboxylase gene for C4 photosynthesis in Zea mays. Mol Gen Genet 224: 325–332 (1990).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Morishima, A. Identification of preferred binding sites of a light-inducible DNA-binding factor (MNF1) within 5′-upstream sequence of C4-type phosphoenolpyruvate carboxylase gene in maize. Plant Mol Biol 38, 633–646 (1998). https://doi.org/10.1023/A:1006085812507
Issue Date:
DOI: https://doi.org/10.1023/A:1006085812507