Abstract
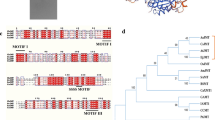
We describe the molecular and functional characterization of three closely related S-adenosyl-L-methionine synthetase (SAMS) isoenzymes from Catharanthus roseus (Madagascar periwinkle). The genes are differentially expressed in cell cultures during growth of the culture and after application of various stresses (elicitor, nutritional down-shift, increased NaCl). Seedlings revealed organ-specific expression and differential gene regulation after salt stress. A relationship analysis indicated that plant SAMS group in two main clusters distinguished by characteristic amino acid exchanges at specific positions, and this suggested differences in the enzyme properties or the regulation. SAMS1 and SAMS2 are of type I and SAMS3 is of type II. The properties of the isoenzymes were compared after heterologous expression of the individual enzymes, but no significant differences were detected in a) optima for temperature (37 to 45 °C) or pH (7 to 8.3); b) dependence on cations (divalent: Mg2+, Mn2+, Co2+; monovalent: K+, \({\text{NH}}_{\text{4}}^{\text{ + }} \), Na+); c) Kms for ATP and L-methionine; d) inhibition by reaction products (S-adenosyl-L-methionine, PPi, Pi), by the reaction intermediate tripolyphosphate, and by the substrate analogues ethionine and cycloleucine; e) response to metabolites from the methyl cycle (L-homocysteine) or from related pathways (L-ornithine, putrescine, spermidine, spermine); f) native protein size (gel permeation chromatography). The results represent the first characterization of plant SAMS isoenzyme properties with individually expressed proteins. The possibility is discussed that the isoenzyme differences reflect specificities in the association with enzymes that use S-adenosyl-L-methionine.
Similar content being viewed by others
References
Aarnes H: Partial purification and characterization of methionine adenosyltransferase from pea seedlings. Plant Sci Letters 10: 381–390 (1977).
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ: Basic local alignment search tool. J Mol Biol 215: 403–410 (1990).
Alvarez L, Mingorance J, Pajares MA, Mato JM: Expression of rat liver S-adenosylmethionine synthetase in Escherichia coli results in two active oligomeric forms. Biochem J 301: 557–561 (1994).
Anderson JW: Sulfur metabolism in plants. In: Miflin BJ, Lea PJ (eds) The Biochemistry of Plants. A Comprehensive Treatise. Vol. 16. pp. 327–381, Academic Press, New York (1990).
Bairoch A: PROSITE: a dictionary of sites and patterns in proteins. Nucleic Acids Res 20: 2013–2018 (1992).
Boerjan W, Bauw G, Van Montagu M, Inzé D: Distinct phenotypes generated by overexpression and suppression of Sadenosyl-L-methionine synthetase reveal developmental patterns of gene silencing in tobacco. Plant Cell 6: 1401–1414 (1994).
Bryan JK: Advances in the biochemistry of amino acid biosynthesis. In: Miflin BJ, Lea PJ (eds) The Biochemistry of Plants. A Comprehensive Treatise. Vol. 16. pp. 161–195, Academic Press, New York (1990).
Chang S, Puryear J, Cairney J: A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Reporter 11: 113–116 (1993).
Cherest H, Surdin-Kerjan Y: The two methionine adenosyltransferases in Saccharomyces cerevisiae: evidence for the existence of dimeric enzymes. Mol Gen Genet 182: 65–69 (1981).
Chiang PK, Cantoni GL: Activation of methionine for transmethylation. Purification of the S-adenosylmethionine synthetase of Baker's yeast and its separation into two forms. J Biol Chem 252: 4506–4513 (1977).
Chou T-C, Talalay P: The mechanism of S-adenosyl-Lmethionine synthesis by purified preparations of Baker's yeast. Biochemistry 11: 1065–1073 (1972).
Crowe J, Henco K: The QIA expressionist. 2nd Edition, Qiagen Inc. Chatsworth, California, USA (1992).
Drummond JT, Jarrett J, González JC, Huang S, Matthews RG: Characterization of non-radioactive assays for cobalamin-dependent and cobalamin-independent methionine synthase enzymes. Analyt Biochem 228: 323–329 (1995).
Eichel J, González JC, Hotze M, Matthews RG, Schröder J: Vitamin B12-independent L-methionine synthase from a higher plant (Catharanthus roseus): molecular characterization, regulation, heterologous expression, and enzyme properties. Eur J Biochem 230: 1053–1058 (1995).
Espartero J, Pintor-Toro JA, Pardo JM: Differential accumulation of S-adenosylmethionine synthetase transcripts in response to salt stress. Plant Mol Biol 25: 217–227 (1994).
Fliegmann J, Schröder G, Schanz S, Britsch L, Schröder J: Molecular analysis of chalcone and dihydropinosylvin synthase from Scots pine (Pinus sylvestris), and differential regulation of these and related enzyme activities in stressed plants. Plant Mol Biol 18: 489–503 (1992).
Gómez-Gómez L, Carrasco P: Hormonal regulation of Sadenosylmethionine synthase transcripts in pea ovaries. Plant Mol Biol 30: 821–832 (1996).
Habereder H, Schröder G, Ebel J: Rapid induction of phenylalanine ammonia-lyase and chalcone synthase mRNAs during fungus infection of soybean (Glycine max L.) roots or elicitor treatment of soybean cell cultures at the onset of phytoalexin synthesis. Planta 177: 58–65 (1989).
Hahlbrock K, Wellmann E: Light-independent induction of enzymes related to phenylpropanoid metabolism in cell suspension cultures from parsley. Biochim Biophys Acta 304: 702–706 (1973).
Higgins DG, Bleasby AJ, Fuchs R: CLUSTAL V: improved software for multiple sequence alignment. Cabios 8: 189–191 (1992).
Kende H: Ethylene biosynthesis. Annu Rev Plant Physiol Plant Mol Biol 44: 283–307 (1993).
Kunkel TA: Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA 82: 488–492 (1985).
Lanz T, Schröder G, Schröder J: Differential regulation of genes for resveratrol synthase in cell cultures of Arachis hypogaea. Planta 181: 169–175 (1990).
Maina CV, Riggs PD, Grandea AGI, Slatko BE, Moran LS, Tagliamonte JA, McReynolds LA, Guan C: A vector to express and purify foreign proteins in Escherichia coli by fusion to, and separation from, maltose binding protein. Gene 74: 365–373 (1988).
Markham GD, Hafner EW, Tabor CW, Tabor H: Sadenosylmethionine synthetase from Escherichia coli. J Biol Chem 255: 9082–9092 (1980).
Masuta C, Tanaka H, Uehara K, Kuwata S, Kowai A, Noma M: Broad resistance to plant viruses in transgenic plants conferred by antisense inhibition of a host gene essential in S-adenosylmethionine-dependent transmethylation reactions. Proc Natl Acad Sci USA 92: 6117–6121 (1995).
Mathur M, Sachar RC: Phytohormonal regulation of Sadenosylmethionine synthetase and S-adenosylmethionine levels in dwarf pea epicotyls. FEBS Lett 287: 113–117 (1991).
Mathur M, Saluja D, Sachar RC: Post-transcriptional regulation of S-adenosylmethionine synthetase fromits stored mRNA in germinated wheat embryos. Biochim Biophys Acta 1078: 161–170 (1991).
Mathur M, Satpathy M, Sachar RC: Phytohormonal regulation of S-adenosylmethionine synthetase by gibberellic acid in wheat aleurones. Biochim Biophys Acta 1137: 338–348 (1992).
Mathur M, Sharma N, Sachar RC: Differential regulation of S-adenosylmethionine synthetase isozymes by gibberellic acid in dwarf pea epicotyls. Biochim Biophys Acta 1162: 283–290 (1993).
Mitsui K, Teraoka H, Tsukada K: Complete purification and immunochemical analysis of S-adenosylmethionine synthetase from bovine brain. J Biol Chem 263: 11211–11216 (1988).
Pajares MA, Corrales F, Durán C, Mato JM, Alvarez L: How is rat liver S-adenosylmethionine synthetase regulated? FEBS Lett 309: 1–4 (1992).
Pajares MA, Durán C, Corrales F, Mato JM: Protein kinase C phosphorylation of rat liver S-adenosylmethionine synthetase: dissociation and production of an active monomer. Biochem J 303: 949–955 (1994).
Peleman J, Boerjan W, Engler G, Seurinck J, Botterman J, Alliotte T, Van Montagu M, Inzé D: Strong cellular preference in the expression of a housekeeping gene of Arabidopsis thaliana encoding S-adenosylmethionine synthetase. Plant Cell 1: 81–93 (1989).
Peleman J, Saito K, Cottyn B, Engler G, Seurinck J, Van Montagu M, Inzé D: Structure and expression analyses of the S-adenosylmethionine synthetase gene family in Arabidopsis thaliana. Gene 84: 359–369 (1989).
Saitou N, Nei M: The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 (1987).
Sakata SF, Shelly LL, Ruppert S, Schutz G, Chou JY: Cloning and expression of murine S-adenosylmethionine synthetase. J Biol Chem 268: 13978–13986 (1993).
Sambrook J, Fritsch EF, Maniatis T: Molecular Cloning: a Laboratory Manual. 2nd Edition, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. (1989).
Schröder G, Beck M, Eichel J, Vetter H-P, Schröder J: HSP90 homolog from periwinkle (Catharanthus roseus): cDNA sequence, regulation of protein expression, and location in the endoplasmic reticulum. Plant Mol Biol 23: 583–594 (1993).
Schröder G, Brown JWS, Schröder J: Molecular analysis of resveratrol synthase: cDNA, genomic clones and relationship with chalcone synthase. Eur J Biochem 172: 161–169 (1988).
Schröder G, Schröder J: cDNAs for S-adenosyl-L-methionine decarboxylase from Catharanthus roseus, heterologous expression, identification of the proenzyme processing site, evidence for the presence of both subunits in the active enzyme, and a conserved region in the 5′ mRNA leader. Eur J Biochem 228: 74–78 (1995).
Schröder G, Waitz A, Hotze M, Schröder J: cDNA for Sadenosyl-L-homocysteine hydrolase from Catharanthus roseus. Plant Physiol 104: 1099–1100 (1994).
Schröder J, Kreuzaler F, Schäfer E, Hahlbrock K: Concomitant induction of phenylalanine ammonia-lyase and flavanone synthase mRNAs in irradiated plant cells. J Biol Chem 254: 57–65 (1979).
Suma Y, Shimizu K, Tsukada K: Isozymes of Sadenosylmethionine synthetase from rat liver: isolation and characterization. J Biochem 100: 67–75 (1986).
Tabor CW, Tabor H: Methionine adenosyltransferase (Sadenosylmethionine synthetase) and S-adenosylmethionine decarboxylase. Adv Enzymol 56: 251–282 (1984).
Takusagawa F, Kamitori S, Misaki S, Markham GD: Crystal structure of S-adenosylmethionine synthetase. J Biol Chem 271: 136–147 (1996).
Thomas D, Surdin-Kerjan Y: SAM1, the structural gene for one of the S-adenosylmethionine synthetases in Saccharomyces cerevisiae. Sequence and expression. J Biol Chem 262: 16704–16709 (1987).
Tiburcio AF, Kaur-Sawhney R, Galston AW: Polyamine metabolism. In: Miflin BJ, Lea PJ (eds) The Biochemistry of Plants. A Comprehensive Treatise. Vol. 16. pp. 283–325, Academic Press, New York (1990).
Tropf S, Lanz T, Rensing SA, Schröder J, Schröder G: Evidence that stilbene synthases have developed from chalcone synthases several times in the course of evolution. J Mol Evol 38: 610–618 (1994).
Van Breusegem F, Dekeyser R, Gielen J, Van Montagu M, Caplan A: Characterization of a S-adenosylmethionine synthetase gene in rice. Plant Physiol 105: 1463–1464 (1994).
Van de Peer Y, De Wachter R: TREECON: a software package for the construction and drawing of evolutionary trees. Comput Applic Biosci 9: 177–182 (1993).
Van Doorsselaere J, Gielen J, Inzé D: A cDNA encoding Sadenosyl-L-methionine synthetase from poplar. Plant Physiol 102: 1365–1366 (1993).
Vetter H-P, Mangold U, Schröder G, Marner F-J, Werck-Reichhart D, Schröder J: Molecular analysis and heterologous expression of an inducible cytochrome P-450 protein from periwinkle (Catharanthus roseus L.). Plant Physiol 100: 998–1007 (1992).
Winters AL, Gallagher J, Pollock CJ, Farrar JF: Isolation of a gene expressed during sucrose accumulation in leaves of Lolium temulentum L. J Exp Botany 46: 1345–1350 (1995).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Schröder, G., Eichel, J., Breinig, S. et al. Three differentially expressed S-adenosylmethionine synthetases from Catharanthus roseus: molecular and functional characterization. Plant Mol Biol 33, 211–222 (1997). https://doi.org/10.1023/A:1005711720930
Issue Date:
DOI: https://doi.org/10.1023/A:1005711720930