Abstract
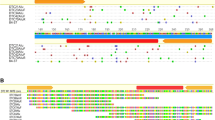
We have characterised from Xenopus laevis two new short interspersed repetitive elements, we have named Glider and Vision, that belong to the family of miniature inverted-repeat transposable elements (MITEs). Glider was first characterised in an intronic region of the α-tropomyosin (α-TM) gene and database search has revealed the presence of this element in 10 other Xenopus laevis genes. Glider elements are about 150 bp long and for some of them, their terminal inverted repeats are flanked by potential target-site duplications. Evidence for the mobility of Glider element has been provided by the presence/absence of one element at corresponding location in duplicated α-TM genes. Vision element has been identified in the promoter region of the cyclin dependant kinase 2 gene (cdk2) where it is boxed in a Glider element. Vision is 284 bp long and is framed by 14-bp terminal inverted repeats that are flanked by 7-bp direct repeats. We have estimated that there are about 20,000 and 300 copies of Glider and Vision respectively scattered throughout the laevis genome. Every MITEs elements but two described in our study are found either in 5′ or in 3′ regulatory regions of genes suggesting a potential role in gene regulation.
Similar content being viewed by others
References
Altschul, S.F., T.L. Madden, A.A. Schäffer, J. Zhang, Z. Zhang, W. Miller & D.J. Lipman, 1997. Gapped Blast and PSI-Blast: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402.
Bisbee, C.A., M.A. Baker, A.C. Wilson, I. Haji-Azimi & M. Fischberg, 1977. Albumin phylogeny for clawed frogs (Xenopus). Science 195: 785–787.
Britten, R.J., 1996. DNA sequence insertion and evolutionary variation in gene regulation. Proc. Natl. Acad. Sci. USA 93: 9374–9377.
Bureau, T.E. & S.R. Wessler, 1992. Tourist: a large family of small inverted repeat elements frequently associated with maize genes. Plant Cell 4: 1283–1294.
Bureau, T.E. & S.R. Wessler, 1994a. Stowaway: a new family of inverted repeat elements associated with the genes of both monocotyledonous and dicotyledonous plants. Plant Cell 6: 907–916.
Bureau, T.E. & S.R. Wessler, 1994b. Mobile inverted-repeat elements of the Tourist family are associated with the genes of many cereal grasses. Proc. Natl. Acad. Sci. USA 91: 1411–1415.
Bureau, T.E., P.C. Ronald & S.R. Wessler, 1996. A computer-based systematic survey reveals the predominance of small invertedrepeat elements in wild-type rice genes. Proc. Natl. Acad. Sci. USA 93: 8524–8529.
Finnegan, D.J., 1989. Eukaryotic transposable elements and genome evolution. Trends Genet. 5: 103–107.
Gaillard, C., N. Thézé, H. Lerivray, S. Hardy, D. Lepetit & P. Thiébaud, 1998a. A novel tropomyosin isoform encoded by the Xenopus laevis α-TM gene is expressed in the brain. Gene 207: 235–239.
Gaillard, C., N. Thézé, S. Hardy, M.R. Allo, E. Ferrasson & P. Thiébaud, 1998b. α tropomyosin gene expression in Xenopus laevis: Differential promoter usage and controlled expression by myogenic factors. Dev. Genes Evol. 207: 435–445.
Gaillard, C., H. Lerivray, N. Thézé, B. Cooper, D. Lepetit, T. Mohun & P. Thiébaud, 1999. Differential expression of two skeletal muscle beta-tropomyosin mRNAs during Xenopus laevis development. Int. J. Dev. Biol. 43: 175–178.
Graf, J.D. & H.R. Kobel, 1991. Genetics of Xenopus laevis. Meth. Cell Biol. 36: 19–34.
Hardy, S. & P. Thiébaud, 1992. Isolation and characterization of cDNA clones encoding the skeletal and smooth muscle Xenopus laevis α tropomyosin isoforms. Biochim. Biophys. Acta 1131: 239–242.
Hardy, S., M. Fiszman, H.B. Osborne & P. Thiébaud, 1991. Characterization of muscle and non muscle Xenopus laevis tropomyosin mRNAs transcribed from the same gene. Eur. J. Biochem. 202: 431–440.
Hardy, S., N. Thézé, D. Lepetit, M.R. Allo & P. Thiébaud, 1995. The Xenopus laevis TM-4 gene encodes non muscle and cardiac tropomyosin isoforms through alternative splicing. Gene 156: 265–270.
Henikoff, S., E.A. Greene, S. Pietrokovski, P. Bork, T.K. Attwood & L. Hood, 1997. Gene families: the taxonomy of protein paralogs and chimeras. Science 278: 609–614.
Higgins, D.G. & S.M. Sharp, 1988. Clustal: a package for performing multiple sequence alignment on a microcomputer. Gene 73: 237–244.
Izsvák, Z., Z. Ivics, N. Shimoda, D. Mohn, H. Okamoto & P.B. Hackett, 1999. Short inverted-repeat transposable elements in teleost fish and implications for a mechanism of their amplification. J. Mol. Evol. 48: 13–21.
Kafatos, F.C., C.W. Jones & A. Efstratiadis, 1979. Determination of nucleic acid sequence homologies and relative concentrations by a dot hybridization procedure. Nucleic Acids Res. 7: 1541–1552.
Kay, B.K. & I.B. Dawid, 1983. The 1723 element: a long, homogeneous, highly repeated DNA unit interspersed in the genome of Xenopus laevis. J. Mol. Biol. 170: 583–596.
Kidwell, M.G. & D. Lisch, 1997. Transposable elements as sources of variation in animals and plants. Proc. Natl. Acad. Sci. USA 94: 7704–7711.
Kobel, H.R. & L. Du Pasquier, 1986. Genetic of polyploid Xenopus. Trends Genet. 2: 310–315.
Lees-Miller, J.P., L.O. Goodwin & D.M. Helfman, 1990. Three novel brain tropomyosin isoforms are expressed from the rat alpha-tropomyosin gene through the use of alternative promoters and alternative RNA processing. Mol. Cell. Biol. 10: 1729–1742.
Lemonnier, M., L. Balvay, V. Mouly, D. Libri & M. Fiszman, 1991. The chicken gene encoding the alpha isoform of tropomyosin of fast-twitch muscle fibers: organization, expression and identification of the major proteins synthesized. Gene 107: 229–240.
Matzke, M.A. & A.J. Matzke, 1998. Epigenetic silencing of plant transgenes as a consequence of diverse cellular defence responses. Cell. Mol. Life Sci. 54: 94–103.
Morgan, G.T. & K.M. Middleton, 1990. Short interspersed repeats from Xenopus that contain multiple octamer motifs are related to known transposable elements. Nucleic Acids Res. 18: 5781–5786.
Olive, M., N. Thézé, M. Philippe, J.P. Le Pennec & H. Lerivray, 1994. Cloning of the Xenopus laevis cdk2 promoter and functional analysis in oocytes and during early development. Gene 151: 81–88.
Pozueta-Romero, J., G. Houlne & R. Schantz, 1996. Nonautonomous inverted repeat Alien transposable elements are associated with genes of both monocotyledonous and dicotyledonous plants. Gene 171: 147–153.
Sambrook, J., E.F. Fritsch & T. Maniatis, 1989. Molecular cloning. A Laboratory Manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
Sanger, F., S. Nicklen & A.R. Coulson, 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74: 5463–5467.
Smit, A.F. & A.D. Riggs, 1996. Tiggers and DNA transposon fossils in the human genome. Proc. Natl. Acad. Sci. USA 93: 1443–1448.
Tu, Z., 1997. Three novel families of miniature inverted-repeat transposable elements are associated with genes of the yellow fever mosquito, Aedes aegypti. Proc. Natl. Acad. Sci. USA 94: 7475–7480.
Ñnsal, K. & G.T. Morgan, 1995. Novel group of families of short interspersed repetitive elements (SINEs) in Xenopus: evidence of a specific target site for DNA-mediated transposition of inverted-repeat SINEs. J. Mol. Biol. 248: 812–823.
Wessler, S.R., T.E. Bureau & S.E. White, 1995. LTR-retrotransposons and MITEs: important players in the evolution of plant genomes. Curr. Opin. Genet. Dev. 5: 814–821.
White, S.E., L.F. Habera & S.R. Wessler, 1994. Retrotransposons in the flanking regions of normal plant genes: a role for copia-like elements in the evolution of gene structure and expression. Proc. Natl. Acad. Sci. USA 91: 11792–11796.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Lepetit, D., Pasquet, S., Olive, M. et al. Glider and Vision: Two New Families of Miniature Inverted-Repeat Transposable Elements in Xenopus Laevis Genome. Genetica 108, 163–169 (2000). https://doi.org/10.1023/A:1004173315419
Issue Date:
DOI: https://doi.org/10.1023/A:1004173315419