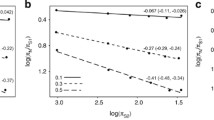
Abstract
The principal methods of using DNA sequence information to test the neutral theory of evolution and polymorphism are described. These include the use of synonymous and nonsynonymous substitutions for detecting purifying and positive selection, the analysis of nucleotide diversity, mismatch analysis and the HKA, McDonald-Kreitman, Tajima and Ewens-Watterson tests. Analysis of the covariation of different kinds of molecular markers and the relationship between genetic variation and fitness is also considered. Examples of the use of these approaches in a wide variety of marine organisms are described. It is emphasised that tests of neutral theory, in addition to providing important fundamental knowledge about the action of evolutionary forces, provide valuable information about the influence of environmental and demographic factors.
Similar content being viewed by others
References
Aguade, M. & C. H. Langley, 1994. Polymorphism and divergence in regions of low recombination in Drosophila. In Golding, B. (ed.), Non-Neutral Evolution: Theories and Data. Chapman & Hall, New York: 67–76.
Arnason, E. & S. Palsson, 1996. Mitochondrial cytochrome b DNA sequence variation of Atlantic cod Gadus morhua, from Norway. Mol. Ecol. 5: 715–724.
Beerli, P., 1998. Estimation of migration rates and populations sizes in geographically structured populations. In Carvalho, G. (ed.), Advances in Molecular Ecology. NATO Science, Series A: Life Sciences. IOS Press, Amsterdam: 39–53.
Begun, C. J. & C. F. Aquadro, 1992. Levels of naturally occurring DNA polymorphism correlate with recombination rates of Drosophila melanogaster. Nature 356: 519–520.
Bertorelle, G. & M. Slatkin, 1995. The number of segregating sites in expanding human populations, with implications for estimates of demographic parameters. Mol. Biol. Evol. 12: 887–892.
Boom, J. D. G., E. G. Boulding & A. T. Beckenback, 1994. Mitochondrial DNA variation in introduced populations of pacific oyster, Crassostrea gigas, in British Columbia. Can. J. Fish. aquat. Sci. 51: 1608–1614.
Britten, H. B., 1996. Meta-analysis of the association between multilocus heterozygosity and fitness. Evolution 50: 2158–2164.
Brookfield, J. F. Y & P. M. Sharp, 1994. Neutralism and selectionism face up to DNA data. Trends Genet. 10: 109–111.
Buroker, N. E., 1983. Population genetics of the American oyster Crassostrea virginica along the Atlantic coast and the Gulf of Mexico. Mar. Biol. 75: 99–112.
Burton, R. S. & B.-N. Lee, 1994. Nuclear and mitochondrial gene genealogies and allozyme polymorphisms across a major phylogeographic break in the copepod Tigriopus californicus. Proc. natn. Acad. Sci. U.S.A. 91: 5197–5201.
Charlesworth, D., B. Charlesworth & M. T. Morgan, 1995. The pattern of neutral molecular variation under the background selection model. Genetics 141: 1605–1617.
Endo, T., K. Ikeo & T. Gojobori, 1996. Large-scale search for genes on which positive selection may operate. Mol. Biol. Evol. 13: 685–690.
Fevolden, S. E. & R. Schneppenheim, 1989. Genetic homogeneity of krill (Euphausia superba Dana) in the Southern Ocean. Polar. Biol. 9: 533–539.
FitzSimmons, N. N., C. Moritz, C. J. Limpus, L. Pope & R. Prince, 1997. Geographic structure of mitochondrial and nuclear gene polymorphisms in Australian green turtle populations and male-biased gene flow. Genetics 147: 184–1854.
Ford, M. J., 1998. Testing models of migration and isolation among populations of chinook salmon (Oncorhynchus tschawytscha). Evolution 52: 539–557.
Fu, Y. X., 1994. A phylogenetic estimator of effective population size or mutation rate. Genetics 136: 685–692.
Gillespie, J. H., 1989. Could natural selection account for molecular evolution and polymorphism? Genome 31: 311–315.
Gillespie, J. H., 1991. The causes of molecular evolution. Oxford University Press, New York.
Gillespie, J. H., 1994. Alternatives to the neutral theory. In Golding, B. (ed.), Non-Neutral Evolution: Theories and Data. Chapman & Hall, New York: 1–17.
Graur, D. & W. H. Li, 1991. Neutral mutation hypothesis test. Nature 354: 114–115.
Hare, M. P. & J. C. Avise, 1998. Population structure in the American oyster as inferred by nuclear gene genealogies. Mol. Biol. Evol. 15: 119–128.
Hare, M. P., S. A. Karl & J. C. Avise, 1996. Anonymous nuclear DNA markers in the American oyster and their implications for the heterozygote deficiency phenomenon in marine bivalves. Mol. Biol. Evol. 13: 334–345.
Hudson, R. R., 1991. Gene genealogies and the coalescent approach. Oxford Surv. evol. Biol. 7: 1–44.
Hudson. R. R., M. Kreitman & M. Aguade, 1987. A test of neutral molecular evolution based on nucleotide data. Genetics 116: 153–159.
Karl, S. A. & J. C. Avise, 1992. Balancing selection at allozyme loci in oysters: implications from nuclear RFLP's. Science 256: 100–102.
Karl, S. A., S. Schultz, D. Desbruyeres, R. Lutz & R. C. Vrijenhoek, 1996. Molecular analysis of gene flow in the hydrothermal vent clam (Calyptogena magnifica). Mol. mar. Biol. Biotech. 5: 193–202.
Kimura, M., 1983. The neutral theory of molecular evolution. Cambridge University Press, London.
Kimura, M. & J. F. Crow, 1964. The number of alleles that can be maintained in a finite population. Genetics 49: 725–738.
Lavery, S., C. Moritz & D. R. Fielder, 1996. Genetic patterns suggest exponential population growth in a declining species. Mol. Biol. Evol. 13: 1106–1113.
Lee, Y.-H. & V. D. Vacquier, 1992. The divergence of species-specific abalone sperm lysins is promoted by positive Darwinian selection. Biol. Bull. 182: 97–104.
Lee, Y.-H., T. Ota & V. D. Vacquier, 1995. Positive selection is a general phenomenon in the evolution of abalone sperm lysin. Mol. Biol. Evol. 12: 231–238.
Lewontin, R. C. & J. Krakauer, 1975. Testing the heterogeneity of F values. Genetics 80: 397–398.
McDonald, J. H., 1994. Detecting natural selection by comparing geographic variation in protein and DNA polymorphisms. In Golding, B. (ed.), Non-Neutral Evolution: Theories and Data. Chapman & Hall, New York: 88–100.
McDonald, J. H. & M. Kreitman, 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351: 652–654.
McDonald, J. H., B. C. Verrelli & L. B. Geyer, 1996. Lack of geographic variation in anonymous nuclear polymorphisms in the American oyster, Crassostrea virginica. Mol. Biol. Evol. 13: 1114–1118.
Metz, C. E., G. Gomez-Gutierrez & V. D. Vacquier, 1998b. Mitochondrial DNA and bindin gene sequence evolution among allopatric species of the sea urchin genus Arbacia. Mol. Biol. Evol. 15: 185–195.
Metz, E. C. & S. R. Palumbi, 1996. Positive selection and sequence rearrangements generate extensive polymorphism in the gamete recognition protein bindin. Mol. Biol. Evol. 13: 397–406.
Metz, C. E., R. Robles-Sikisaka & V. D. Vacquier, 1998a. Nonsynonymous substitution in abalone sperm fertilization genes exceeds substitution in introns and mitochondrial DNA. Proc. natn. Acad. Sci. U.S.A. 95: 10676–10681.
Moriyama, E. N. & J. R. Powell, 1996. Intraspecific nuclear DNA variation in Drosophila. Mol. Biol. Evol. 13: 261–277.
Nei, M. & T. Maruyama, 1975. Lewontin-Krakauer test for neutral genes. Genetics 80: 395.
Ohta, T., 1992. The nearly neutral theory of molecular evolution. Ann. Rev. Ecol. Syst. 23: 263–286.
Pogson, G. H. & S. E. Fevolden, 1998. DNA heterozygosity and growth rate in the Atlantic cod Gadus morhua (L). Evolution 52: 915–920.
Pogson, G. H., K. A. Mesa & R. G. Boutilier, 1995. Genetic population structure and gene flow in the Atlantic cod Gadus morhua: a comparison of allozyme and nuclear RFLP loci. Genetics 139: 375–385.
Pogson, G. H. & E. Zouros, 1994. Allozyme and RFLP heterozygosities as correlates of growth rate in the scallop Placopecten magellanicus: a test of the associative overdominance hypothesis. Genetics 137: 221–231.
Powers, D. A. & P. M. Schulte, 1998. Evolutionary adaptations of gene structure and expression in natural populations in relation to a changing environment: a multidisciplinary approach to address the million-year saga of small fish. J. exp. Zool. 282: 71–94.
Quesada, H., M. Warren & D. O. F. Skibinski, 1998. Nonneutral evolution and differential mutation rate of gender-associated mitochondrial DNA lineages in the marine mussel Mytilus. Genetics 149: 1511–1526.
Quesada, H., R. Wenne & D. O. F. Skibinski, 1999. Interspecies transfer of female mitochondrial DNA is coupled with role-reversals and departure from neutrality in the mussel Mytilus trossulus. Mol. Biol. Evol. 16: 655–665.
Rand, D. M., 1996. Neutrality tests of molecular markers and the connection between DNA polymorphism, demography and conservation biology. Conserv. Biol. 10: 665–671.
Rand, D. M. & L. M. Kann, 1998. Mutation and selection at silent and replacement sites in the evolution of animal mitochondrial DNA. Genetica 102/103: 393–407.
Raybould, A. F., R. J. Mogg & R. T. Clarke, 1996. The genetic structure of Beta vulgaris ssp. maritima (sea beet) populations: RFLPs and isozymes show different patterns of gene flow. Heredity 77: 245–250.
Raybould, A. F, R. J. Mogg, C. Aldam, C. J. Gliddon, R. S. Thorpe & R. T. Clarke, 1998. The genetic structure of sea beet (Beta vulgaris ssp. maritima) populations. III. Detection of isolation by distance at microsatellite loci. Heredity 80: 127–132.
Reeb, C. A. & J. C. Avise, 1990. A genetic discontinuity in a continuously distributed species: mitochondrial DNA in the American oyster, Crassostrea virginica. Genetics 124: 397–406.
Robertson, A., 1975. Remarks on the Lewontin-Krakauer test. Genetics 80: 396.
Rogers, A. R., 1995. Genetic evidence for a Pleistocene population explosion. Evolution 49: 608–615.
Rogers, A. R. & H. Harpending, 1992. Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 9: 552–569.
Slatkin, M., 1995. A measure of population subdivision based on microsatellite allele frequencies. Genetics 139: 457–462.
Slatkin, M. & R. R. Hudson, 1991. Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics 129: 555–562.
Stewart, D. T., E. R. Kenchington, R. K. Singh & E. Zouros, 1996. Degree of selective constraint as an explanation of the different rates of evolution of gender-specific mitochondrial DNA lineages in the mussel Mytilus. Genetics 143: 1349–1357.
Stewart, D. T., C. Saavedra, R. R. Stanwood, A. O. Ball & E. Zouros, 1995. Male and female mitochondrial DNA lineages in the blue mussel (Mytilus edulis) species group. Mol. Biol. Evol. 12: 735–747.
Swanson, W. J. & V. D. Vacquier, 1998. Concerted evolution in an egg receptor for a rapidly evolving abalone sperm protein. Science 281: 710–712.
Tajima, F., 1983. Evolutionary relationship of DNA sequences in finite populations. Genetics 105: 437–460.
Tajima, F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595.
Taylor, M. F. J., Y. Shen & M. E. Kreitman, 1995. A population genetic test of selection at the molecular level. Science 270: 1497–1499.
Vacquier, V. D., W. J. Swanson & Y.-H. Lee, 1997. Positive Darwinian selection on two homologous fertilization proteins: what is the selective pressure driving their divergence? J. mol. Evol. 44: S15–S22.
Watterson, G. A., 1975. On the number of segregation sites. Theor. Pop. Biol. 7: 256–276.
Watterson, G. A., 1978. The homozygosity test of neutrality. Genetics 88: 405–417.
Whitlock, M. C. & D. E. McCauley, 1999. Indirect measures of gene flow and migration. F ST ≠ 1/(4N m + 1). Heredity 82: 117–125.
Whittam, T. S. & M. Nei, 1991. Neutral mutation hypothesis test. Nature 354: 115–116.
Williams, S. T. & J. A. H. Benzie, 1997. Indo-West Pacific patterns of genetic differentiation in the high-dispersal starfish Linckia laevigata. Mol. Ecol. 6: 559–573.
Zane, I., L. Ostellari, L. Maccatrozzo, L. Bargelloni, B. Battaglia & T. Patarnello, 1998. Molecular evidence for genetic subdivision of Antarctic krill (Euphausia superba Dana) populations. Proc. r. Soc., Lond. B 265: 2387–2391.
Zouros, E. & G. H. Pogson, 1994. The present status of the relationship between heterozygosity and heterosis. In Beaumont, A. R. (ed.), Genetics and the Evolution of Aquatic Animals. Chapman and Hall, London: 135–146.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Skibinski, D.O.F. DNA tests of neutral theory: applications in marine genetics. Hydrobiologia 420, 137–152 (2000). https://doi.org/10.1023/A:1003954229804
Issue Date:
DOI: https://doi.org/10.1023/A:1003954229804