Abstract
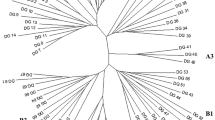
DNA amplification fingerprinting (DAF) with arbitrary oligonucleotide primers was used to study genetic relationships between cultivars of flowering dogwood (Cornus florida L.), evaluate extent of plant hybridization, and generate markers in pseudo-testcross mapping at the intraspecific level. Modified Taguchi optimization methods defined a robust DAF system based on high annealing temperature (48–52 °C) and primer concentration (typically 8 μM) that was used to study genetic diversity of representative dogwood cultivars and hybrids. Phenetic analysis using cluster and numerical methods showed that: (1) cultivars were relatively conserved at the genetic level; (2) their hybridization could be identified in the F1 progeny in the absence of phenotypic or physiological markers; (3) several cultivars grouped according to their recorded ancestry; and (4) dogwood anthracnose-resistant lines originally selected in Catoctin Mountain Park (Maryland) grouped separately from those of southern origin. The DAF protocol was also tested in pseudo-testcross mapping of dogwood at the intraspecific level. A preliminary screening of parents ‘Pink Sachet’ and ‘Fragrant Cloud’ and 7 F1 segregants with 22 octamer primers produced 703 amplified loci, 30 and 39 of which were male and female markers segregating at 1:1 ratios with 98.6% confidence levels in pseudo-testcross configuration. Overall results show that DAF generated markers very efficiently (3 per primer) despite the close relatedness of parental dogwood cultivars. This study constitutes the basis for a future genetic linkage mapping and marker-assisted selection (MAS) effort initially targeted to control important fungal diseases in dogwood.
Similar content being viewed by others
References
Bassam, B.J. & G. Caetano-Anollés, 1993. Silver staining of DNA in polyacrylamide gels. Appl Biochem Biotechnol 42: 181–188.
Bassam, B.J., G. Caetano-Anollés & P.M. Gresshoff, 1991. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem 196: 80–83.
Bassam, B.J., G. Caetano-Anollés & P.M. Gresshoff, 1992. DNA amplification fingerprinting of bacteria. Appl Microbiol Biotechnol 38: 70–76.
Bentley, S. & B.J. Bassam, 1996. A robust DNA amplification fingerprinting system applied to analysis of genetic variation within Fusarium oxysporum f.sp. cubense. J Phytopathology 144: 207–213.
Caetano-Anollés, G., 1996. Scanning of nucleic acids by in vitro amplification: New developments and applications. Nature biotechnology 14: 1668–1674.
Caetano-Anollés, G., 1998. DAF optimization using Taguchi methods and the effect of thermal cycling parameters on DNA amplification. Biotechniques 25: 472–480.
Caetano-Anollés, G. & R.N. Trigiano, 1997. Nucleic acid markers in agricultural biotechnology. AgBiotech News & Information 9: 235–242.
Caetano-Anollés, G., B.J. Bassam & P.M. Gresshoff, 1991. DNA amplification fingerprinting using short arbitrary oligonucleotide primers. Bio/technology 9: 553–557.
Caetano-Anollés, G., B.J. Bassam & P.M. Gresshoff, 1992. Primertemplate interactions during DNA amplification fingerprinting with single arbitrary oligonucleotides. Mol Gen Genet 235: 157–165.
Caetano-Anollés, G., L.M. Callahan, P.E. Williams, K.R. Weaver & P.M. Gresshoff, 1995. DNA amplification fingerprinting analysis of bermudagrass (Cynodon): genetic relationships between species and interspecific crosses. Theor Appl Genet 91: 228–235.
Cai, Q., C.L. Guy & G.A. Moore, 1994. Extension of the linkage map in Citrus using random amplified polymorphic DNA (RAPD) markers and RFLP mapping of cold-acclimatationresponsive loci. Theor Appl Genet 89: 606–614.
Cerny, T.A., G. Caetano-Anollés, R.N. Trigiano & T.W. Starman, 1996. Molecular phylogeny and DNA amplification fingerprinting of Petunia taxa. Theor Appl Genet 92: 1009–1016.
Cobb, B.D. & J.M. Clarkson, 1994. A simple procedure for optimising the polymerase chain reaction (PCR) using modified Taguchi methods. Nucleic Acids Res 22: 3801–3805.
Daughtrey, M. & C.R. Hibben, 1994. Dogwood anthracnose: a new disease threatens two native Cornus species. Ann Rev Phytopathol 32: 61–73.
Eyde, R.H., 1988. Comprehending Cornus: puzzles and progress in the systematics of the dogwoods. Bot Rev 54: 233–251.
Graham, E.T., R.N. Trigiano & M.T. Windham, 1995. Evaluation of potential applications of Catoctin Mountain dogwoods. Tenn Agri Sci 175: 38–44.
Grattapaglia, D. & R.R. Sederoff, 1994. Genetic linkage maps of Eucalyptus grandis and E. urophylla using a pseudo-testcross mapping strategy and RAPD markers. Genetics 137: 1121–1137.
He, G., C.S. Prakash & R.L. Jarret, 1995. Analysis of genetic diversity in a sweetpotato (Ipomoaea batatas) germplasm collection using DNA amplification fingerprinting. Genome 38: 938–945.
Hemmat, M., N.F. Weeden, A.G. Manganaris & D.M. Lawson, 1994. Molecular-marker linkage map for apple. J Heredity 85: 4–11.
Jaccard, P., 1908. Nouvelles recherches sur la distribution florale. Bull Soc Vaud Sci Nat 44: 223–270.
Kubisiak, T.L., C.D. Nelson, W.L. Nance & M. Stine, 1995. RAPD linkage mapping in a longleaf pine × slash pine F1 family. Theo Appl Genet 90: 1119–1127.
Penner, G.A., A. Bush, R. Wise, W. Kim, L. Domier, K. Kasha, A. Laroche, G. Scoles, S.J. Molnar & G. Fedal, 1993. Reproducibility of random amplified polymorphic DNA (RAPD) analysis among laboratories. PCR Methods Applic 2: 341–345.
Rohlf, F.J., 1992. Numerical taxonomy and multivariate analysis system (NTSYS-pc), version 1.8. Exeter Software, Setauket, NY.
Qiu, J., E. van Santen & S. Tuzun, 1995. Optimization of DNA amplification fingerprinting techniques to study genetic relationships of white lupin germplasm. Plant Breeding 114: 525–529.
Ritter, E., C. Gebhardt & F. Salamini, 1990. Estimation of recombination frequencies and construction of linkage maps from crosses between heterozygous parents. Genetics 125: 645–654.
Santamour, F.S. Jr & A.J. McArdle, 1985. Cultivar checklists of the large-bracted dogwoods: Cornus florida, C. kousa and C. nuttalli. J Arboric 11: 29–36.
Scott, M.C., G. Caetano-Anollés & R.N. Trigiano, 1996. DNA amplification fingerprinting identifies closely related Chrysanthemum cultivars. J Amer Soc Hort Sci 121: 1043–1048.
Taguchi, G., 1987. System of experimental design. UNIPUB/Kraus International Publishers, White Plains, NY.
Trigiano, R.N. & M.T. Windham, 1997. Are ‘Barton’ and ‘Cloud Nine’ the same cultivar? Proc Southern Nurserymen's Assoc Res Conf 42: 182–184.
Weaver, K.R., L.M. Callahan, G. Caetano-Anollés & P.M. Gresshoff, 1995. DNA amplification fingerprinting and hybridization analysis of centipedegrass. Crop Sci 35: 881–885.
Williams, J.G.K., A.R. Kubelik, K.J. Livak, J.A. Rafalski & S.V. Tingey, 1990. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 18: 6531–6535.
Witte, W.T., 1995. Dogwood culture in nursery and landscape. Tenn Agri Sci 175: 47–51.
Wolff, K., E.D. Schien & J. Peters-van Rijn, 1993. Optimizing the generation of random amplified polymorphic DNAs in chrysanthemum. Theor Appl Genet 86: 1033–1037.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Caetano-anollés, G., Schlarbaum, S. & Trigiano, R. DNA amplification fingerprinting and marker screening for pseudo-testcross mapping of flowering dogwood ( Cornus florida L.). Euphytica 106, 209–222 (1999). https://doi.org/10.1023/A:1003568604537
Issue Date:
DOI: https://doi.org/10.1023/A:1003568604537