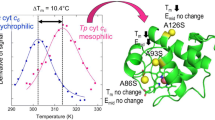
Abstract
Fitch and Markowitz' theory of concomitantly variable codons (covarions) in evolution predicted the existence of functional correlation in amino acid residue mutations among present-day cytochromes c. Mutational analysis was carried out on yeast iso-2-cytochrome c, where hydrophobic core residues I20, M64, L85, and M98 and surface residue L9 were mutated, in selected combinations, to those found in mammalian and bird cytochromes c. The functionality assay is based upon the ability of yeast cells to grow in YPGE medium. Furthermore, experiments on the single M64L and M98L mutations as well as the double M64L/M98L mutation using NMR showed that the effects of these mutations are to perturb the structural integrity of the protein. We identified functional correlation in two cases of a pair of residue mutations, the I20 → V and M98 → L pair and the L9 → I and L85 → I pair. In both cases, only one of the two alternative, putative evolutionary pathways leads to a functional protein and the corresponding pairs of residue mutations are among those found in present-day cytochromes c. Since valine is predicted to be at position 20 in the ancestral form of cytochrome c, the present data provide an explanation for the ancient requirement of leucine rather than methionine in position 98. The present data provide further evidence for the role of those specific atom–atom interactions in directing a pathway in the evolutionary changes of the amino acid sequence that have taken place in cytochrome c, in accordance with Fitch and Markowitz.
Similar content being viewed by others
REFERENCES
Amegazie, B., Zitomer, R., and Hollenberg, C. P. (1990). Characterization of cytochrome c gene from the starch-fermenting yeast Schwanniomyces occidentalis and its expression in baker's yeast. Yeast 6:429.
Arcangioli, B., and Lescure, B. (1985). Identification of proteins involved in the regulation of yeast iso-2-cytochrome expression by oxygen. EMBO J. 4:2627.
Auld, D. S., and Pielak, G. J. (1991). Constraints on amino acid substitutions in the N-terminal helix of cytochrome c explored by random mutagenesis. Biochemistry 30:8684.
Baba, M. L., Darga, L. L., Goodman, M., and Czelusniak, J. (1981). Evolution of cytochrome c investigated by the maximum parsimony method. J.Mol.Evol. 17:197.
Bax, A., and Davis, D. G. (1985). M1EV-17-based two dimensional homonuclear magnetization transfer spectroscopy. J.Magn.Reson. 65:355.
Bushnell, G. W., Louie, G. V., and Brayer, G. D. (1990). High-resolution three-dimensional structure of horse heart cytochrome c. J.Mol.Biol. 214:585.
Cutler, R. L., Pielak, G. J., and Smith, M. (1987). Replacement of cysteine-107 of Saccharomyces cerevisiae iso-1-cytochrome c with threonine: Improved stability of the mutant protein. Protein Eng. 1:95.
Das, G., Hickey, D. R., McLeudon, D., McLeudon, G., and Sherman, F. (1989). Dramatic thermostabilization of yeast iso-1cytochrome c by an asparagine 3 isoleucine replacement at position 57. Proc.Natl.Acad.Sci.USA 86:496.
Dickerson, R. E., Takano, T., Eisenberg, D., Kallai, O. B., Samson, L., Cooper, A., and Margoliash, E. (1971). Ferri cytochrome c. I. General features of the horse and bonito proteins at 2.8 A resolution. J.Biol.Chem. 246:1511.
Downie, J. A., Stewart, J. W., Brockman, N., Schweingruber, A. M., and Sherman, F. (1977). Structural gene for yeast iso-2-cytochrome c. J.Mol.Biol. 113:369.
Faye, G., Leung, D. W., Tatchell, K., Hall, B. D., and Smith, M. (1981). Deletion mapping sequences essential for in vitro transcription of the iso-1-cytochrome c gene. Proc.Natl.Acad.Sci.USA 78:2258.
Fitch, W. M. (1976). The molecular evolution of cytochrome c in eukaryotes. J.Mol.Evol. 8:13.
Fitch, W. M., and Margoliash, E. (1970). The usefulness of amino acid and nucleotide sequences in evolutionary studies. In Steere, W. C., Dobzhansky, T., and Hect, M. K., (eds.), Evolutionary Biology, Vol. IV, Appleton–Century–Crofts, New York, pp. 67–109.
Fitch, W. M., and Markowitz, E. (1970). An improved method for determining codon variability in a gene and its application to the rate of fixation of mutations in evolution. Biochem.Genet. 4:579.
Frauenfelder, H., Sligar, S. G., and Wolynes, P. G. (1991). The energy landscapes and motions of proteins. Science 254:1587.
Frederick, Z. L., and Pielak, G. J. (1993). Exploring the interface between the N-and C-terminal helices of cytochrome c by random mutagenesis within C-terminal helix. Biochemistry 32:929.
Guarente, L. (1984). Yeast promoters: Positive and negative elements. Cell 36:799.
Hampsey, D. H., Das, G., and Sherman, F. (1986). Amino acid replacements in yeast iso-1-cytochromre c: Comparison with the phylogenetic series and the tertiary structure of related cytochromes c. J.Biol.Chem. 261:3259.
Hennig, B. (1975). Change of cytochrome c structure during development of the mouse. Eur. J.Biochem. 55:167.
Hickey, D. R., McLeudon, G., and Sherman, F. (1988). Thermodynamic stabilities of yeast iso-1-cytochromes c having amino acid substitution for lysine 32. J.Biol.Chem. 263:18298.
Inoue, S., Hiroyoshi, T., Matsubara, H., and Yamanaka, T. (1984). Complete amino-acid sequences of two isocytochromes c of the housefly, Musca domestica L., and their developmental variation in different tissues. Biochim.Biophys.Acta 790:188.
Janbon, G., Rustchenko, E. P., Klug, S., Scherer, S., and Sherman, F. (1977). Phylogenetic relationships of fungal cytochromes c. Yeast 13:985.
Jeener, J., Meier, B. H., Bachman, P., and Ernst, R. R. (1979). Investigation of exchange processes by two-dimensional spectroscopy. J.Chem.Phys. 71:4546.
Kaminsky, L. S., Miller, V. J., and Davison, A. J. (1973). Thermodynamic studies of the opening of the heme crevice of ferricytochrome c. Biochemistry 12:2215.
Kunkel, T. A. (1985). Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc.Natl.Acad.Sci.USA 82:488.
Koppenol, W. H., and Margoliash, E. (1982). The asymmetric distribution of charges on the surface of horse cytochrome c. J.Biol.Chem. 257:4426.
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680.
Lederer, F., Simon, A. M., and Verdiere, J. (1972). Saccharomyces cerevisiae iso-cytochromes c: Revision of the amino acid sequence between the cysteine residues. Biochem.Biophys.Res. Commun. 47:55.
Limbach, K. J., and Wu, R. (1985). Characterization of two Drosophila melanogaster cytochrome c genes and their transcripts. Nucleic Acids Res. 13:631.
Lyddiart, A., Peacock, D., and Boutler, D. (1978). Evolutionary change in invertebrate cytochrome c. J.Mol.Evol. 11:35.
Margoliash, E. (1972). The molecular variations of cytochrome c as a function of the evolution of species. Harvey Lect. 66:177.
Margoliash, E., Smith, E. L., Kriel, G., and Tuppy, H. (1961). Amino acid sequence of horse-heart cytochrome c. Nature 192:1121.
Margoliash, E., Fitch, W. M., Markowitz, E., and Dickerson, R. E. (1972). Functional limits of cytochrome c variability. In: Ehrenberg, A. (ed.), Oxidation-Reduction Enzymes, Winksol, Stockholm, pp. 5–17.
Matton, J. R., and Sherman, F. (1966). Reconstitution of phosphorylating electron transport in mitochondria from a cytochromre c-deficient yeast mutant. J.Biol.Chem. 241:4330.
McGee, W. A., Rosell, F. I., Liggins, J. R., Rodriguez-Ghidarpour, S., Luo, Y., Chen, J., Brayer, G. D., Mauk, A. G., and Nall, B. T. (1996). Thermodynamic cycles as probes of structure in unfolded proteins. Biochemistry 35:1995.
Montgomery, D. L., Leung, D. W., Smith, M., Shalit, P., Faye, G., and Hull, B. D. (1980). Isolation and sequence of the gene for iso-2 cytochrome c in Saccharomyces serevisiae. Proc.Natl.Acad. Sci.USA 77:54.
Moore, G. F., and Pettigrew, G. W. (1990). Cytochrome c: Evolutionary, Structural and Physicochemical Aspects, Springer-Verlag, Berlin.
Murphy, M. E. P., Nall, B. T., and Brayer, G. D. (1992). Structure determination and analysis of yeast iso-2-cytochrome c and a composite mutant protein. J.Mol.Biol. 227:160.
Nall, B. T., and Landers, T. A. (1981). Guanidine hydrochloride induced unfolding of yeast iso-2 cytochrome c. Biochemistry 20:5403.
Narita, K., and Titani, K. (1968). The amino acid sequence of cytochrome c from Candida krusei. J.Biochem. 63:226.
Narita, K., and Titani, K. (1969). The complete amino acid sequence in baker's yeast cytochrome c. J.Biochem. 65:259.
Nolan, C., Weiss, L. J., Adams, J. J., and Margoliash, E. (1968). Unpublished results cited in Dickerson, R. E., and Timkovich, R. (1975). Cytochrome c. In Boyer, P. D. (ed.), The Enzymes, Academic Press, Orlando, FL, p. 397.
Parr, G. R., Hantgan, R. R., and Taniuchi, H. (1978). Formation of two alternative complementing structure from a cytochrome c heme fragment (residues 1 to 38) and the apoprotein. J.Biol. Chem. 253:5381.
Pelletier, H., and Kraut, J. (1992). Crystal structure of a complex between electron transfer partners, cytochrome c peroxidase and cytochrome c. Science 258:1748.
Pettigrew, G. W., and Moore, G. R. (1987). Cytochrome c: Biological Aspects, Springer-Verlag, Berlin.
Picos, M. A. F., Torres, A. M. R., Ramil, E., Cerdan, M. E., Brennig, K. D. Hollenberg, C. P., and Zitomer, R. S. (1993). Sequence of a cytochrome c gene from Kluyveromyces lactis and its upstream region. Yeast 9:201.
Rance, M., Sprensen, O. W., Bedenhausen, G., Wagner, G., Ernst, R. R., and Wuthrich, K. (1983). Improved spectral resolution in cosy IH NMR spectra of proteins via double quantum filtering. Biochem.Biophys.Res.Commun. 117:479.
Rizzo, P., Tinello, C., Punturieri, A., and Taniuchi, H. (1992). A study of hydrogen exchange of monoclonal antibodies: Specificity of the antigen-binding induced conformational stabilization. Biochim.Biophys.Acta 1159:169.
Shechter, E., and Saludjian, P. (1967). Conformation of ferricytchrome c. IV. Relationship between optical absorption and protein conformation. Biopolymer 5:788.
Sherman, F., and Stewart, J. W. (1971). Genetics and biosynthesis of cytochrome c. Annu.Rev. Genet. 5:257.
Sherman, F., Stewart, J. W., Margoliash, E., Parker, J., and Campbell, W. (1966). The structural gene for yeast cytochrome c. Proc.Natl.Acad.Sci.USA 55:1498.
States, D. J., Haberkorn, R. A., and Ruber, D. J. (1982). A two-dimensional nuclear Overhanser experiment with pure absorption phase in four quadrants. J.Magn.Reson. 48:286.
Sugeno, K., Narita, K., and Titani, K. (1971). The amino acid sequence of cytochrome c from Debaryomyces kloeckeri. J.Biochem. 70:659.
Takano, T., Kallai, O. B., Swanson, R., and Dickerson, R. E. (1973). The structure of ferrocytochrome c at 2.45 A resolution. J.Biol.Chem. 248:5234.
Tarr, G. E., and Fitch, W. M. (1976). Amino acid sequence of cytochrome c from Tetrahymena pyriformis phenoset A. Biochem.J. 159:193.
Turcott, B., and Guarente, L. (1992). HAP1 positive control mutants specific for one of two binding sites. Genes Dev. 6:2001.
Varadarajan, R., Connely, R. R., Sturtevant, J. M., and Richards, F. M. (1992). Heat capacity changes for protein-peptide interactions in the ribonuclease S system. Biochemistry 31:1421.
Williams, G., Moore, G. R., and Williams, R. J. P. (1985). Bilogical electron transfer: the structure, dynamics and reactivity of cytochrome c. Comments Inorg.Chem. IV:55.
Wood, L. C., Muthukrishnan, K., White, T. B., Ramdas, L., and Nall, B. T. (1988). Construction and characterization of mutant iso-2-cytochromes c with replacement of conserved prolines. Biochemistry 27:8554.
Yaoi, Y. (1967). Comparison of the primary structures of cytochromes c from wild and respirationdeficient mutant yeasts. J.Biochem. 61:54.
Zitomer, R. S., and Lowry, C. V. (1992). Regulation of gene expression by oxygen in Saccharomyces cerevisiae. Microbiol.Rev. 56:1.
Zuckerkandl, E. (1963). Perspectives in molecular anthropology. In Washburn, S. L. (ed.), Classi-fication and Human Evolution, Aldine, Chicago, pp. 243–272.
Zuckerkandl, E. (1976). Evolutionary processes and evolutionary noise at the molecular level. J.Mol. Evol. 7:167.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Fisher, A., Shi, Y., Ritter, A. et al. Functional Correlation in Amino Acid Residue Mutations of Yeast Iso-2-Cytochrome c that Is Consistent with the Prediction of the Concomitantly Variable Codon Theory in Cytochrome c Evolution. Biochem Genet 38, 177–196 (2000). https://doi.org/10.1023/A:1001977630789
Issue Date:
DOI: https://doi.org/10.1023/A:1001977630789