Abstract
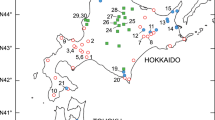
We investigated the levels and patterns of genetic diversity of Pinus densata Master in Yunnan. Horizontal starch-gel electrophoresis was performed on macrogametophytes collected from nine populations in northwestern Yunnan, China. Compared with other gymnosperm species, P. densata has higher mean values for all measures of genetic diversity. Allozyme polymorphism (0.99 criterion) was 97.0% and 71.4% at the species and population levels, respectively. The average number of alleles per locus was 3.1 and 2.0 at the species and population levels. Mean expected heterozygosity was substantially higher in P. densata than average values investigated for other gymnosperms both at the population (H ep = 0.174±0.031) and at the species (H es = 0.190) levels. Of the total genetic variation, less than 12% was partitioned among populations (G ST = 0.112). Our allozyme survey supports the suggestion that the observed higher diversity in P. densata may be attributed partly to its hybrid origin between two genetically distinct species, P. yunnanensis and P. tabulaeformis. In addition, we suggest that introgression would give rise to the increase in genetic diversity occurring in P. densata.
Similar content being viewed by others
REFERENCES
Boyle, T. J. B. and Morgenstern, E. K. (1987). Some aspects of the population structure of black spruce in central New Brunswick. Sil.Genet. 36:53.
Cheng, W. C., and Fu, L. K. (1978). Flora Reipublicae Popularis Sinicae, Science Press, Beijing, Vol. 7, pp. 255–259.
Farjon, A. (1984). Pines: Drawings and Descriptions of the Genus, E. Brill, Leiden, The Netherlands.
Ge, S., Wang, H. Q., Zhang, C. M., and Hong, D. Y. (1997). Genetic diversity and population differentiation of Cathaya argyrophylla in Bamian Mountain. Acta Bot.Sinica 39:266.
Ge, S., Hong, D. Y., Wang, H. Q., Liu, Z. Y., and Zhang, C. M. (1998). Population genetic structure and conservation of an endangered conifer, Cathaya argyrophylla (Pinaceae). Int.J.Plant Sci. 159:351.
Guan, C. T. (1981). Fundamental features of the distribution of Coniferae in Sichuan. Acta Phytotaxon Sinica 11:393.
Hamrick, J. L., Godt, M. J. W., and Sherman-Broyles, S. L. (1992). Factors influencing levels of genetic diversity in woody plant species. New Forests 6:95.
Mirov, N. T. (1967). The Genus Pinus, Ronald Press, New York.
Nei, M. (1973). Analysis of gene diversity in subdivided populations. Proc.Natl.Acad.Sci.USA 70:3321.
Nei, M. (1978). Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583.
Soltis, D. E., Haufler, C. H., Darrow, D. C., and Gastony, G. J. (1983). Starch gel electrophoresis of ferns: A compilation of grinding buffers, gel and electrode buffers, and staining schedules. Am. Fern J. 73:9.
Swofford, D. L., and Selander, R. B. (1989). BIOSYS-1: A Computer Program for the Analysis of Allelic Variation in Population Genetics and Biochemical Systematics, Release 1.7, Illinois Natural History Survey, Champaign.
Wang, X. R., Szmidt, A. E., Lewandowski, A., and Wang, Z. R. (1990). Evolutionary analysis of Pinus densata Masters, a putative Tertiary hybrid. 1. Allozyme variation. Theor.Appl.Genet. 80:635.
Wang, X. R., and Szmidt, A. E. (1990). Evolutionary analysis of Pinus densata Masters, a putative Tertiary hybrid. 2. A study using species-specific chloroplast DNA markers. Theor.Appl.Genet. 80:641.
Wendel, J. F., and Weeden, N. F. (1989). Visualization and interpretation of plant isozymes. In Soltis, D. E., and Soltis, P. S. (eds.), Isozymes in Plant Biology, Dioscorides Press, Portland, OR, pp. 5–45.
Wheeler, N. C., and Guries, R. P. (1987). A quantitative measure of the introgression between lodgepole and jack pines. Can.J.Bot. 65:1876.
Wright, S. (1978). Evolution and the Genetics of Populations, Vol 4.Variability Within and Among Natural Populations, University of Chicago Press, Chicago.
Wu, C. L. (1956). The taxonomic revision and phytogeographical study of Chinese pines. Acta Phytotaxon Sinica 5:131.
Yu, H. (1996). A Study on Genetic Diversity and Evolution of Pinus yunnanensis Franch, Ph.D. dissertation, Yunnan University, Kunming.
Yu, H., Qian, W., and Huang, R. F. (1999). Allozyme method in study on population genetics of Pinus yunnanensis Franch. Acta Bot.Yunnanica 21:68.
Yu, H., Ge, S., Huang, R. F., and Jiang, H. Q. (2000). A preliminary study on genetic variation and close relationship of Pinus yunnanensis and its closely relative species. Acta Bot.Sinica 42:107.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Yu, H., Ge, S. & Hong, Dy. Allozyme Diversity and Population Genetic Structure of Pinus densata Master in Northwestern Yunnan, China. Biochem Genet 38, 138–146 (2000). https://doi.org/10.1023/A:1001969328972
Issue Date:
DOI: https://doi.org/10.1023/A:1001969328972