Abstract
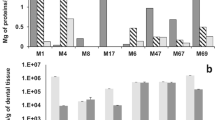
Applications of reliable DNA extraction and amplification techniques to postmortem samples are critical to ancient DNA research. Commonly used methods for isolating DNA from ancient material were tested and compared using both soft tissue and bones from fossil and contemporary museum proboscideans. DNAs isolated using three principal methods served as templates in subsequent PCR amplifications, and the PCR products were directly sequenced. Authentication of the ancient origin of obtained nucleotide sequences was established by demonstrating reproducibility under a blind testing system and by phylogenetic analysis. Our results indicate that ancient samples may respond differently to extraction buffers or purification procedures, and no single method was universally successful. A CTAB buffer method, modified from plant DNA extraction protocols, was found to have the highest success rate. Nested PCR was shown to be a reliable approach to amplify ancient DNA templates that failed in primary amplification.
Similar content being viewed by others
REFERENCES
Barnard, G. F., Puder, M., Begum, N. A., and Chen, L. B. (1994). PCR product sequencing with [α-33P] and [α-32P] dATP. BioTechniques 16:572.
Cano, R. J., and Poinar, H. N. (1993). Rapid isolation of DNA from fossil and museum specimens suitable for PCR. BioTechniques 15:432.
Cattano, C., Smillie, D. M., Gelsthorpe, K., Piccinini, A., Gelsthorpe, A. R., and Sokol, R. J. (1995). A simple method for extracting DNA from old skeletal material. Forens. Sci. Instr. 74:167.
Cooper, A. (1994). DNA from museum specimens. In Herrmann, B., and Hummel, S. (eds.), Ancient DNA, Springer-Verlag, New York, pp. 149–165.
Cooper, A., Mourer-Chauvire, C., Chambers, G. K., von Haeseler, A., Wilson, A. C., and Pääbo, S. (1992). Independent origins of New Zealand moas and kiwis. Proc. Natl. Acad. Sci. USA 89:8741.
Doran, G. H., Dickel, D. N., Ballinger, W. E., Jr., Agee, O. F., Laipis, P. J., and Hauswirth, W. W. (1986). 8000 year old human brain tissue: anatomical cellular and molecular analysis. Nature 323:803.
Doyle, J. J., and Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19:11.
Felsenstein, J. (1990). PHYLIP (Phylogeny Inference Package), Version 3.3, Computer program distributed by University of Washington, Seattle, WA.
Gaensslen, R. E., and Berka, K. (1994). Studies on DNA polymorphisms in human bone and soft tissues. Anal. Chem. Acta 288:3.
Golenberg, E. M. (1994). Fossil samples: DNA from plant compression fossils. In Herrmann, B., and Hummel, S. (eds.), Ancient DNA, Springer Verlag, New York, pp. 233–252.
Golenberg, E. M., Bickel, A., and Wiehs, P. (1996). Effect of highly fragmented DNA on PCR. Nucleic Acids Res. 24:5026.
Hagelberg, E. (1994). Mitochondrial DNA from Ancient bones. In Herrmann, B., and Hummel, S. (eds.), Ancient DNA, Springer-Verlag, New York, pp. 195–204.
Handt, O., Richards, M., Trommsdorff, M., Kilger, C., Simanainen, J., Georgiev, O., Bauer, K., Stone, A., Hedges, R., Schaffner, W., Utermann, G., Sykes, B., and Pääbo, S. (1994). Molecular genetic analyses of the Tyrolean Ice Man. Science 264:1775.
Hänni, C., Laudet, V., Stehelin, D., and Taberlet, P. (1994). Tracking the origins of the cave bear (Ursus spelaeus) by mitochondrial DNA sequencing. Proc. Natl. Acad. Sci. USA 91:12336.
Herrmann, B., and Hummel, S. (eds.) (1994). Ancient DNA, Springer-Verlag, New York.
Höss, M., and Pääbo, S. (1993). DNA extraction from Pleistocene bones by a silica-based purification method. Nucleic Acids Res. 21:3913.
Höss, M., Dilling, A., Currant, A., and Pääbo, S. (1996). Molecular phylogeny of the extinct ground sloth Mylodon darwinii. Proc. Natl. Acad. Sci. USA 93:181.
Hummel, S., Nordsiek, G., and Herrmann, B. (1992). Improved efficiency in amplification of ancient DNA and its sequence analysis. Naturwissenschaften 79:359.
Irwin, D. M., Kocher, T. D., and Wilson, A. C. (1991). Evolution of the cytochrome b gene of mammals. J. Mol. Evol. 32:128.
Janczewski, D. N., Yuhki, N., Gilbert, D. A., Jefferson, G. T., and O'Brien, S. J. (1992). Molecular phylogenetic inference from saber-toothed cat fossils of Rancho La Brea. Proc. Natl. Acad. Sci. USA 89:9769.
Krajewski, C., Driskell, A. C., Baverstock, P. R., and Braun, M. J. (1992). Phylogenetic relationships of the thylacine (Mammalia:Thylacinidae) among dasyuroid marsupials: Evidence from cytochrome b DNA sequences. Proc. R. Soc. Lond. B 250:19.
Lowenstein, J. M., and Shoshani, J. (1996). Proboscidean relationships based on immunological data. In Shoshani, J., and Tassy, P. (eds.), The Proboscidea: Evolution and Palaeoecology of Elephants and Their Relatives, Oxford University Press, Oxford, pp. 49–54.
Pääbo, S. (1989). Ancient DNA: Extraction, characterization, molecular cloning, and enzymatic amplification. Proc. Natl. Acad. Sci. USA 86:1939.
Pääbo, S., Gifford, J. A., and Wilson, A. C. (1988). Mitochondrial DNA sequences from a 7000-year old brain. Nucleic Acids Res. 16:9775.
Pääbo, S., Higuchi, R. G., and Wilson, A. C. (1989). Ancient DNA and the polymerase chain reaction. J. Biol. Chem. 264:9709.
Rogers, S. O. (1994). Phylogenetic and taxonomic information from herbarium and mummified DNA. In Adams, R. P., Miller, J. S., Golenberg, E. M., and Adams, J. E. (eds.), Conservation of Plant Genes II: Utilization of Ancient and Modern DNA, Missouri Botanical Garden, pp. 47–67.
Salo, W. L., Aufderheide, A. C., Buikstra, J., and Holcomb, T. (1994). Identification of Mycobacterium tuberculosis DNA in a pre-Columbian Peruvian mummy. Proc. Natl. Acad. Sci. USA 91:2091.
Shoshani, J. (1996). Para-or monophyly of the gomphotheres and their position within Proboscidea. In Shoshani, J., and Tassy, P. (eds.), The Proboscidea: Evolution and Palaeoecology of Elephants and Their Relatives, Oxford University Press, Oxford, pp. 149–177.
Shoshani, J., et al. (1982). On the dissection of a female Asian elephant (Elephas maximus maximus Linnaeus, 1758) and data from other elephants. Elephant 2:3.
Shoshani, J., Lowenstein, J. M., Walz, D. A., and Goodman, M. (1985). Proboscidean origins of mastodon and woolly mammoth demonstrated immunologically. Paleobiology 11:429.
Shoshani, J., Fisher, D. C., Zawiskie, J. M., Thurlow, S. J., Shoshani, S. L., Benninghoff, W. S., and Zoch, F. H. (1989). The Shelton Mastodon Site: Multidisciplinary study of a late Pleistocene (Twocreekan) locality in southeastern Michigan. Contrib. Mus. Paleontol. Univ. Mich. 27:393.
Swofford, D. L. (1991). PAUP: Phylogenetic Analysis Using Parsimony, Version 3.0, Computer program distributed by the Illinois Natural History Survey, Champaign, IL.
Tassy, P. (1996). Who is who among the Proboscidea. In Shoshani, J., and Tassy, P. (eds.), The Proboscidea: Evolution and Palaeoecology of Elephants and Their Relatives, Oxford University Press, Oxford. pp. 39–48.
Thomas, R. H. (1994). Molecules, museums and vouchers. Trends Ecol. Evol. 9:413.
Tuross, N. (1994). The biochemistry of ancient DNA in bone. Experientia 50:530.
Vochot, A.-M., and Monnerot, M. (1996). Extraction, amplification and sequencing of DNA from formaldehyde-fixed specimens. Ancient Biomolecules 1:3.
Wayne, R. K., and Jenks, S. M. (1991). Mitochondrial DNA analysis implying extensive hybridization of the endangered red wolf Canis rufus. Nature 351:565.
Yang, H., Golenberg, E. M., and Shoshani, J. (1996). Phylogenetic resolution within Elephantidae using fossil DNA sequence from American mastodon (Mammut americanum) as an outgroup. Proc. Natl. Acad. Sci. USA 93:1190.
Yang, H., Golenberg, E. M., and Shoshani, J. (1997). A blind testing design for authenticating ancient DNA sequences. Mol. Phylogenies Evol. (in press).
Zimmermann, K., Pischinger, K., and Mannhalter, J. W. (1994). Nested primer PCR detection limits of HIV-1 in the background of increasing numbers of lysed cells. BioTechniques 17:18.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Yang, H., Golenberg, E.M. & Shoshani, J. Proboscidean DNA from Museum and Fossil Specimens: An Assessment of Ancient DNA Extraction and Amplification Techniques. Biochem Genet 35, 165–179 (1997). https://doi.org/10.1023/A:1021902125382
Issue Date:
DOI: https://doi.org/10.1023/A:1021902125382