Abstract
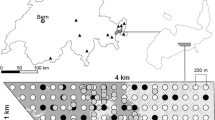
Non-invasive genetic sampling (NGS) is being increasingly applied in wildlife monitoring and population genetic research. This study was designed to evaluate the use of NGS for reconstructing the spatial structure of populations of large felids. We developed a procedure for reliably genotyping individuals of Eurasian lynx (Lynx lynx) from samples obtained through a hair-trapping scheme based on a network of lynx scent-marking sites. The spatial locations of the identified genotypes were matched with the home ranges distribution of radio-tracked individuals, thus cross-checking the accuracy of the two methods. We analyzed DNA extracted from 170 hair samples and 11 blood samples from live-trapped lynx collected in 2004–2009 in the Białowieża Primeval Forest, Poland. We obtained PCR products in 96 (67%) hair samples; 82 (85%) of them were reliably genotyped at 12 autosomal microsatellite loci following a multiple-tubes protocol and stringent quality-controls of the data set. The sample included 29 distinct genotypes; 18 were found only in hair samples, five were determined only in live-trapped animals, and six in both hair and blood samples. Based on linkage disequilibrium we estimated an effective population size Ne = 20.3 (90% CI = 15–28). The total population size estimated with Capwire was Nc =32 (95% CI = 25–37) in close agreement with the observed number of genotypes. The genotypes obtained from hair samples were re-sampled on average 3.9 times and 50% of them were recorded for more than one year. The spatial distribution of six hair-genotypes was consistent with their home ranges obtained by radio-tracking in the same period. The distribution ranges of hair-trapped genotypes overlapped on average in 86.4% (mode 100%) with home ranges of the corresponding individuals. Hair-trapping and molecular identification is a reliable method for reconstructing the spatial organization of lynx population. It is likely to be also efficiently used in other rare and endangered species of felids in combination with data from other monitoring techniques, such as radio- and snow-tracking and photo-trapping.
Similar content being viewed by others
References
Belkhir, K., Borsa, P., Chikhi, L., Raufaste, N., Bonhomme, F., 1996–2004. GENETIX 4.05, logiciel sous Windows TM pour la génétique des populations. Laboratoire Génome, Populations, Interactions, CNRS UMR 5000, Université de Montpellier II, Montpellier, France (in French).
Boulanger, J., Himmer, S., Swan, C., 2004. Monitoring of grizzly bear population trends and demography using DNA mark-recapture methods in the Owikeno Lake area of British Columbia. Can. J. Zool. 82, 1267–1277.
Boulanger, J., Proctor, M., Himmer, S., Stenhouse, G., Paetkau, D., Cranston, J., 2006. An empirical test of DNA mark-recapture sampling strategies for grizzly bear. Ursus 17, 149–158.
Breitenmoser, U., 1989. A footsnare for medium-sized Carnivores. Cat News 11, 20.
Breitenmoser, U., Breitenmoser-Würsten, C., Okarma, H., Kaphegyi, T., Kaphegyi-Wallmann, U., Müller, U.M., 2000. Action Plan for the Conservation of the Eurasian Lynx in Europe (Lynx lynx). Nature and Environment 112. Council of Europe, Strasbourg Cedex, France.
Breitenmoser-Würsten, C., Vandel, J.M., Zimmermann, F., Breitenmoser, U., 2007. Demography of lynx Lynx lynx in the Jura Mountains. Wildl. Biol. 13, 381–392.
Broquet, T., Petit, E., 2004. Quantifying genotyping errors in noninvasive population genetics. Mol. Ecol. 13, 3601–3608.
Castro-Arellano, I., Madrid-Luna, C., Lacher Jr., T.E., Leon-Paniagua, L., 2008. Hair-trap efficacy for detecting mammalian carnivoresin the tropics. J. Wildl. Manage. 72, 1405–1412.
Crow, J.F., Kimura, M., 1970. An Introduction to Population Genetics Theory. Harper & Row, New York, NY, USA.
Davison, A., Birks, J.D.S., Brookes, R.C., Braithwaite, T.C., Messanger, J.E., 2002. On the origin of faeces: morphological versus molecular methods for surveying rare carnivores from their scats. J. Zool. (Lond.) 257, 141–143.
De Barba, M., Waits, L.P., Garton, E.O., Genovesi, P., Randi, E., Mustoni, A., Groff, C., 2010. The power of genetic monitoring for studying demography, ecology and genetics of a reintroduced brown bear population. Mol. Ecol. 19, 3938–3951.
Delibes, M., Rodríguez, A., Ferreras, P., 2000. Action Plan for the conservation of the Iberian lynx (Lynx pardinus) in Europe. Council of Europe, Strasbourg, France.
Downey, P.J., Hellgren, E.C., Caso, A., Carvajal, S., Frangioso, K., 2007. Hair snares for noninvasive sampling of felids in North America: do gray foxes affect success. J. Wildl. Manage. 71, 2090–2094.
Eizirik, E., Kim, J.H., Menotti-Raymond, M., Crawshaw, P.G., O’Brien, S.J., Johnson, W.E., 2001. Phylogeography, population history and conservation genetics of jaguars (Panthera onca, Mammalia, Felidae). Mol. Ecol. 10, 65–79.
Fabbri, E., Miquel, C., Lucchini, V., Santini, A., Caniglia, R., Duchamp, C., Weber, J.M., Lequette, B., Marucco, F., Boitani, L., Fumagalli, L., Taberlet, P., Randi, E., 2007. From the Appennines to the Alps: colonization genetics of the naturally expanding Italian wolfs (Canis lupus) population. Mol. Ecol. 16, 1661–1671.
Faliński, J.B., 1986. Vegetation Dynamics in Temperate Lowland Primeval Forest. Dr. W. Junk Publishers, Dordrecht, Germany.
Gerloff, U., Schlötterer, C., Rassmann, K., Rambold, I., Hohmann, G., Fruth, B., Tautz, D., 1995. Amplification of hypervariable simple sequence repeats (microsatellites) from excremental DNA of wild living bonobos (Pan paniscus). Mol. Ecol. 4, 515–518.
Gervasi, V., Ciucci, P., Davoli, F., Boulanger, J., Boitani, L., Randi, E., 2010. Addressing challenges in non invasive capture–recapture based estimates of small populations: a pilot study on the Apennine brown bear. Conserv.Genet. 11, 2299–2310.
Haag, T., Santos, A.S., De Angelo, C., Srbek-Araujo, A.C., Sana, D.A., Morato, R.G., Salzano, F.M., Eizirik, E., 2009. Development and testingofanoptimized method for DNA-based identification of jaguar (Panthera onca) and puma (Puma concolor) faecal samples for use in ecological and genetic studies. Genetics 136, 505–512.
Janečka, J.E., Blankenship, T.L., Hirth, D.H., Tewes, M.E., Kilpatrick, C.W., Grassman Jr., L.I., 2006. Kinship and social structure of bobcats (Lynx rufus) inferred from microsatellite and radio-telemetry data. J. Zool. 269, 494–501.
Jędrzejewski, W., Jędrzejewska, B., Okarma, H., Schmidt, K., Bunevich, A.N., Miłkowski, L., 1996. Population dynamics (1869–1994), demography and home ranges of the lynx in Białowieża Primeval Forest (Poland and Belaruss). Ecography 19, 122–138.
Kalinowski, S.T., Wagner, A.P., Taper, M.L., 2006a. ML-Relate: a computer program for maximum likelihood estimation of relatedness and relationship. Mol. Ecol. Notes 6, 576–579.
Kalinowski, S.T., Sawaya, M., Taper, M.L., 2006b. Individual identification and distributions of genotypic differences. J. Wildl. Manage. 70, 148–150.
Kolbe, J.A., Squires, J.R., Parker, T.W., 2003. An effective box trap for capturing lynx. Wildl. Soc. Bull. 31, 980–985.
McDaniel, G.W., McKelvey, K.S., Squires, J.R., Ruggiero, L.F., 2000. Efficacy of lures and hair snares to detect lynx. Wildl. Soc. Bull. 28, 119–123.
McKelvey, K.S., Von Kienast, J., Aubry, K.B., Koehler, G.M., Maletzke, B.T., Squires, J.R., Lindquist, E.L., Loch, S., Schwartz, M.K., 2006. DNA analysis of hair and scat collected along snow tracks to document the presence of Canada lynx. Wildl. Soc. Bull. 34, 451–455.
Mellen, J.D., 1993. A comparative analysis of scent-marking, social and reproductive behavior in 20 species of small cats (Felis). Am. Zool. 33, 151–166.
Menotti-Raymond, M., David, V.A., Lyons, L.A., Schafer, A.A., Tomlin, J.F., Hutton, M.K., O’Brien, S.J., 1999. A genetic linkage map of microsatellites in the domestic cat (Felis catus). Genomics 57, 9–23.
Miller, C.R., Joyce, P., Waits, L.P., 2002. Assessing allelic dropout and genotyping reliability using maximum likelihood. Genetics 160, 357–366.
Miller, C.R., Joyce, P., Waits, L.P., 2005. A new method for estimating the size of small populations from genetic mark—recapture data. Mol. Ecol. 14, 1991–2005.
Mills, L.S., Citta, J.J., Lair, K.P., Schwartz, M.K., Tallmon, D.A., 2000. Estimating animal abundance using noninvasive DNA sampling: promise and pitfalls. Ecol. Appl. 10, 283–294.
Mowat, G., Paetkau, D., 2002. Estimating marten Martes americana population size using hair capture and genetic tagging. Wildl. Biol. 8 (3), 201–209.
Nichols, J.D., Williams, B.K., 2006. Monitoring for conservation. Trends Ecol. Evol. 21, 668–673.
Nikolov, I.S., Gum, B., Markov, G., Kuehn, R., 2009. Population genetic structure of wild boar Sus scrofa in Bulgaria as revealed by microsatellite analysis. Acta Theriol. 54 (3), 193–205.
Paetkau, D., 2003. Anempirical exploration of data qualityin DNA-based population inventories. Mol. Ecol. 12, 1375–1387.
Palomares, F., Godoy, J.A., Piriz, A., O’Brien, S.J., Johnson, W.E., 2002. Faecal genetic analysis to determine the presence and distribution of elusive carnivores: design and feasibility for the Iberian lynx. Mol. Ecol. 11, 2171–2182.
Peakall, R., Smouse, P.E., 2006. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 6, 288–295.
Pilgrim, K.L., McKelvey, K.S., Riddle, A.E., Schwartz, M.K., 2005. Felid sex identification based on noninvasive genetic samples. Mol. Ecol. Notes 5, 60–61.
Pilot, M., Jędrzejewski, W., Branicki, W., Sidorovich, V.E., Jedrzejewska, B., Stachura, K., Funk, S.M., 2006. Ecological factors influence population genetic structure of European grey wolves. Mol. Ecol. 15, 4533–4553.
Pompanon, F., Bonin, A., Bellemain, E., Taberlet, P., 2005. Genotyping errors: causes, consequences and solutions. Nat. Rev. Genet. 6, 847–859.
Roon, D.A., Thomas, M.E., Kendall, K.C., Waits, L.P., 2005. Evaluating mixed samples as a source of error in non-invasive genetic studies using microsatellites. Mol. Ecol. 14, 195–201.
Rowcliffe, J.M., Carbone, C., 2008. Surveys using camera traps: are we looking to a brighter future? Anim. Conserv. 11, 185–186.
Ruell, E.W., Crooks, K.R., 2007. Evaluation of non-invasive genetic sampling methods for felid and canid populations. J. Wildl. Manage. 71, 1690–1694.
Rueness, E.K., Jorde, P.E., Hellborg, L., Stenseth, N.C., Ellegren, H., Jakobsen, K.S., 2003. Cryptic population structure in a large, mobile mammalian predator: the Scandinavian lynx. Mol. Ecol. 12, 2623–2633.
Sawaya, M.A., Ruth, T.K., Creel, S., Rotella, J.J., Stetz, J.B., Quigley, H.B., Kalinowski, S.T., 2011. Evaluating noninvasive genetic sampling methods for cougars in Yellowstone National Park. J. Wildl. Manage. 75, 612–622.
Schmidt, K., 2008. Behavioural and spatial adaptation of the Eurasian lynx to a decline in prey availability. Acta Theriol. 53, 1–16.
Schmidt, K., Je˛drzejewski, W., Okarma, H., 1997. Spatial organization and social relations in the Eurasian lynx population in Białowieża Primeval Forest. Poland. Acta Theriol. 42, 289–312.
Schmidt, K., Kowalczyk, R., 2006. Using scent-marking stations to collect hair samples to monitor Eurasian lynx populations. Wildl. Soc. Bull. 34, 462–466.
Schmidt, K., Kowalczyk, R., Ozolins, J., Männil, P., Fickel, J., 2009. Genetic structure of the Eurasian lynx population in north-eastern Poland and the Baltic states. Conserv. Genet. 10, 497–501.
Schwartz, M.K., Mills, L.S., Ortega, Y., Ruggiero, L.F., Allendorf, F.W., 2003. Landscape location affects genetic variation of Canada lynx (Lynx canadensis). Mol. Ecol. 12, 1807–1816.
Schwartz, M.K., Luikart, G., Waples, R.S., 2007. Genetic monitoring as a promising tool for conservation and management. Trends Ecol. Evol. 22, 25–33.
Sutherland, W.J., 1996. Ecological Census Techniques: AHandbook. Cambridge University Press, Cambridge, United Kingdom.
Taberlet, P., Griffin, S., Goossens, B., Questiau, S., Manceau, V., Escaravage, N., Waits, L.P., Bouvet, J., 1996. Reliable genotyping of samples with very low DNA quantities using PCR. Nucleic Acids Res. 26, 3189–3194.
Taberlet, P., Waits, L.P., Luikart, G., 1999. Non-invasive genetic sampling: look before you leap. Trends Ecol. Evol. 14, 323–327.
Valière, N., 2002. GIMLET: a computer program for analysing genetic individual identification data. Mol. Ecol. Notes 2, 377–379.
Waits, L.P., Paetkau, D., 2005. Non-invasive genetic sampling tools for wildlife biologists: a review of applications and recommendations for accurate data collection. J. Wildl. Manage. 69 (4), 1419–1433.
Waples, R.S., 2006. A bias correction for estimates of effective population size based on linkage disequilibrium at unlinked gene loci. Conserv. Genet. 7, 167–184.
Wasser, S.K., Houston, C.S., Koehler, G.M., Cadd, G.G., Fain, S.R., 1997. Techinques for the application of faecal DNA methods to field studies of Ursids. Mol. Ecol. 6, 1091–1097.
Weaver, J.L., Wood, P., Paetkau, D., Laack, L.L., 2005. Use of scented hair snares to detect ocelots. Wildl. Soc. Bull. 33 (4), 1384–1391.
Weir, B.S., Cockerham, C.C., 1984. Estimating F-statistics for the analysis of population structure. Evolution 38, 1358–1370.
Woods, J.G., Paetkau, D., Lewis, D., McLellan, B.N., Proctor, M., Strobeck, C., 1999. Genetic tagging of free-ranging black and brown bears. Wildl. Soc. Bull. 27 (3), 616–627.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Davoli, F., Schmidt, K., Kowalczyk, R. et al. Hair snaring and molecular genetic identification for reconstructing the spatial structure of Eurasian lynx populations. Mamm Biol 78, 118–126 (2013). https://doi.org/10.1016/j.mambio.2012.06.003
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1016/j.mambio.2012.06.003