Abstract
Study Design
To test for rare genetic mutations, a cohort of patients with unexplained early-onset scoliosis (EOS) was screened using high-density microarray genotyping. A cohort of patients with adolescent idiopathic scoliosis (AIS) was similarly screened and the results were compared.
Summary of Background Data
Patients with scoliosis in infancy or early childhood (EOS) are at high risk for progressive deformity and associated problems including respiratory compromise. Early-onset scoliosis is frequently associated with genetic disorders but many patients present with nonspecific clinical features and without an associated diagnosis. The authors hypothesized that EOS in these patients may be caused by rare genetic mutations detectable by next-generation genomic methods.
Methods
The researchers identified 24 patients with unexplained EOS from pediatric orthopedic clinics. They genotyped them, along with 39 connecting family members, using the Illumina OmniExpress-12, version 1.0 beadchip. Resulting genotypes were analyzed for chro¬mosomal changes, specifically copy number variation and absence of heterozygosity. They screened 482 adolescent idiopathic scoliosis (AIS) patients and 744 healthy controls, who were similarly genotyped with the same beadchip, for chromosomal changes identified in the EOS cohort.
Results
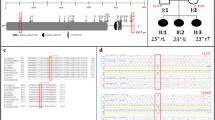
Copy number variation and absence of heterozygosity analyses revealed a genetic diagnosis of chromosome 15q24 microdeletion syndrome in 1 patient and maternal uniparental disomy of chromosome 14 in a second one. Prior genetic testing and clinical evaluations had been negative in both cases. A large novel chromosome 10 deletion was likely causal in a third EOS patient. These mutations identified in the EOS patients were absent in AIS patients and controls, and thus were not associated with AIS or found in asymptomatic individuals.
Conclusions
These data underscore the usefulness of updated genetic evaluations including high-density microarray-based genotyping and other next-generation methods in patients with unexplained EOS, even when prior genetic studies were negative. These data also suggest the intriguing possibility that other mutations detectable by whole genome sequencing, as well as epigenetic effects, await dis¬covery in the EOS population.
Similar content being viewed by others
References
Phillips JH, Knapp Jr DR, Herrera-Soto J. Mortality and morbidity in early-onset scoliosis surgery. Spine (Phila Pa 1976) 2013;38:324–7.
Mortality rate, under-5 (per 1,000 live births). http://data.worldbank.org/indicator/SH.DYN.MORT. Accessed January 30, 2013.
Herring JA. Tachdjian’s pediatric orthopaedics. Philadelphia, PA: WB Saunders; 2008.
Karol LA. Early definitive spinal fusion in young children: what we have learned. Clin Orthop Relat Res 2011;469:1323–9.
Campbell RM. VEPTR expansion thoracoplasty. In: Akbarnia BA, Yazici M, Thompson GH, editors. The growing spine: management of spinal disorders in young children. New York, NY: Springer-Verlag; 2011. p. 469–86.
Akbarnia BA, Mundis Jr GM, Salari P, et al. Dual growing rods. In: Akbarnia BA, Yazici M, Thompson GH, editors. The growing spine: management of spinal disorders in young children. New York, NY: Springer-Verlag; 2010. p. 449-68.
Son-Hing GHTJP, Akbarnia BA, Thompson GH, et al. Single growing rods. In: Akbarnia BA, Yazici M, Thompson GH, editors. The growing spine: management of spinal disorders in young children. New York, NY: Springer-Verlag; 2010. p. 441–8.
Sparrow DB, Chapman G, Smith AJ, et al. A mechanism for gene-environment interaction in the etiology of congenital scoliosis. Cell 2012;149:295–306.
Herring J, editor. Tachdjian’s pediatric orthopaedics, Vol. 1. Philadelphia, PA: WB Saunders; 2008.
Akbarnia BA. Management themes in early onset scoliosis. J Bone Joint Surg Am 2007;89(suppl 1):42–54.
Wynne-Davies R. Infantile idiopathic scoliosis: causative factors, particularly in the first Six months of life. J Bone Joint Surg Br 1975;57:138–41.
Paria N, Copley LA, Herring JA, et al. The impact of large-scale genomic methods in orthopaedic disorders: insights from genome-wide association studies. J Bone J Surg Am 2014;96(38):1–10.
Slavotinek AM. Novel microdeletion syndromes detected by chromosome microarrays. J Hum Genet 2008;124:1–17.
Jiang Y, Tsai TF, Bressler J, Beaudet AL. Imprinting in Angelman and Prader-Willi syndromes. Curr Opin Genet Dev 1998;8:334–42.
Miller DT, Adam MP, Aradhya S, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet 2010;86:749–64.
McQuillan R, Leutenegger AL, Abdel-Rahman R, et al. Runs of homozygosity in European populations. Am J Hum Genet 2008;83:359–72.
Cushman LJ, Torres-Martinez W, Cherry AM, et al. A report of three patients with an interstitial deletion of chromosome 15q24. Am J Med Genet A 2005;137:65–71.
Temple IK, Cockwell A, Hassold T, et al. Maternal uniparental disomy for chromosome 14. J Med Genet 1991;28:511–4.
Temple IK, Shrubb V, Lever M, et al. Isolated imprinting mutation of the DLK1/GTL2 locus associated with a clinical presentation of maternal uniparental disomy of chromosome 14. J Med Genet 2007;44:637–40.
Kagami M, Sekita Y, Nishimura G, et al. Deletions and epimutations affecting the human 14q32.2 imprinted region in individuals with paternal and maternal upd(14)-like phenotypes. Nat Genet 2008;40:237–42.
Hosoki K, Kagami M, Tanaka T, et al. Maternal uniparental disomy 14 syndrome demonstrates Prader-Willi syndrome-like phenotype. J. Pediatr 2009;155:900–3.
Jen JC, Chan WM, Bosley TM, et al. Mutations in a human ROBO gene disrupt hindbrain axon pathway crossing and morphogenesis. Science 2004;304:1509–13.
Sharp AJ, Selzer RR, Veltman JA, et al. Characterization of a recurrent 15q24 microdeletion syndrome. Hum Mol Genet 2007;16:567–72.
Klopocki E, Graul-Neumann LM, Grieben U, et al. A further case of the recurrent 15q24 microdeletion syndrome, detected by array CGH. Eur J Pediatr 2008;167:903–8.
El-Hattab AW, Smolarek TA, Walker ME, et al. Redefined genomic architecture in 15q24 directed by patient deletion/duplication break-point mapping. Hum Genet 2009;126:589–602.
Masurel-Paulet A, Callier P, Thauvin-Robinet C, et al. Multiple cysts of the corpus callosum and psychomotor delay in a patient with a 3.1 Mb 15q24.1q24.2 interstitial deletion identified by array-CGH. Am J Med Genet A 2009;149:1504–10.
Van Esch H, Backx L, Pijkels E, et al. Congenital diaphragmatic hernia is part of the new 15q24 microdeletion syndrome. Eur J Med Genet 2009;52:153–6.
Andrieux J, Dubourg C, Rio M, et al. Genotype-phenotype correlation in four 15q24 deleted patients identified by array-CGH. Am J Med Genet A 2009;149:2813–9.
Sharma S, Gao X, Londono D, et al. Genome-wide association studies of adolescent idiopathic scoliosis suggest candidate susceptibility genes. Hum Mol Genet 2011;20:1456–66.
Takahashi Y, Kou I, Takahashi A, et al. A genome-wide association study identifies common variants near LBX1 associated with adolescent idiopathic scoliosis. Nat Genet 2011;43:1237–40.
Ogata T, Kagami M, Ferguson-Smith AC. Molecular mechanisms regulating phenotypic outcome in paternal and maternal uniparental disomy for chromosome 14. Epigenetics 2008;3:181–7.
Bagri A, Marin O, Plump AS, et al. Slit proteins prevent midline crossing and determine the dorsoventral position of major axonal pathways in the mammalian forebrain. Neuron 2002;33:233–48.
Marillat V, Sabatier C, Failli V, et al. The slit receptor Rig-1/Robo3 controls midline crossing by hindbrain precerebellar neurons and axons. Neuron 2004;43:69–79.
Shashi V, McConkie-Rosell A, Rosell B, et al. The utility of the traditional medical genetics diagnostic evaluation in the context of next-generation sequencing for undiagnosed genetic disorders. Genet Med 2014;16:176–82.
Author information
Authors and Affiliations
Corresponding author
Additional information
Author disclosures: XG (none); GG (none); KR (grant from Fondation Cotrel); CJ (personal fees from Medtronic; patents); SS (none); CAW (none).
Rights and permissions
About this article
Cite this article
Gao, X., Gotway, G., Rathjen, K. et al. Genomic Analyses of Patients With Unexplained Early-Onset Scoliosis. Spine Deform 2, 324–332 (2014). https://doi.org/10.1016/j.jspd.2014.04.014
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1016/j.jspd.2014.04.014