Abstract
Knowledge Graph (KG) has been proven effective in representing and modeling structured information, especially in the medical domain. However, obtaining structured medical information usually depends on the manual processing of medical experts. Meanwhile, the construction of Medical Knowledge Graph (MKG) remains a crucial problem in medical informatization. This work presents a novel method for constructing MKGto drive the application of Rational Drug Use (RDU). We first collect and preprocess the corpora from various types of resources, and then develop a medical ontology via studying the concepts in RDUdomain, authoritative books and drug instructions. Based on the medical ontology, we formulate a scheme to annotate the corpora and construct the dataset for extracting entities and relations. We utilize two mechanisms to extract entities and relations respectively. The former is based on deep learning, while the latter is the rule-based method. In the last stage, we disambiguate and standardize the results of entity relation extraction to construct and enrich the MKG. The experimental results verify the effectiveness of the proposed methods.
Similar content being viewed by others
1 Introduction
With the rapid developments of Internet and Healthcare, construction of intelligent systems for supervising medical processes has become an urgent problem. The systems involve all aspects of medical treatment. Among them, RDUis the most fundamental aspect for implementing medical insurance cost controlling and medical case monitoring. RDUaims at reducing the harmful effects of drugs and ensuring the medication safety of patients under the guidance of modern medicine and pharmaceutical knowledge.
MKGhas become the basic component for building intelligent medical systems. The recent MKGresearches [1,2,3,4,5] only focus on disease and ignore other types of concepts and relations, so that cannot support all demands of RDU. To solve this problem, MKGneeds to involve the concepts such as drugs, diseases, symptoms, patients, etc, as well as the relations between them. In addition, there are many types and different sources of literature in the medical field. It is particularly difficult to integrate multi-source medical corpora, extract the knowledge related to RDUand construct MKGas the database for supporting the system. This paper analyzes the domain characteristics of the corpora, splits the requirements of RDUand constructs an MKGto support the application of RDU.
Contributions This work makes the following contributions:
-
Develop a domain ontology of RDUvia combining the general medical ontology and the specific domain knowledge required for RDU;
-
Formulate an annotation scheme of entity and relation according to the domain ontology, then build a dataset for named entity recognition and relation extraction;
-
Implement a named entity recognition model based on lexicon information and propose a relation extraction model that integrating entity category and character position, then disambiguate and align the extracted triples to construct MKG.
2 Concepts
Knowledge Graph A knowledge graph stores information in the form of triples <subject, predicate, object>. It can be stored as a RDF database of such triples, or equivalently as a graph with nodes and edges.
Domain Ontology A domain ontology is a representation of some part of reality (e.g. medicine, social reality, physics).
Drug The articles intend for use in the prevention, treatment or diagnosis of human diseases, or intend to effect the purposive regulation of human physiological functions, for which indications or major functions, usage and dosage are prescribed, including Chinese medicine, chemical pharmaceuticals, and biological products [6].
Pharmaceutical Ingredient All pharmacological ingredients contained in the drug that are closely related to the clinical application purpose.
Disease Abnormal life activity process of human body due to self stable regulation disorder under the damaging effect of certain reasons.
Symptom Abnormal subjective feeling or some objective pathological changes of the patient caused by a series of abnormal changes in function, metabolism and morphological structure in the process of disease.
Usage and Dosage The specific method, dose and frequency of using one or more drugs for a certain population.
Medication Result The impact on patients under a specific usage and dosage.
Content of RDU Drug interaction, drug compatibility, usage and dosage, medication for special population, adverse drug reactions, allergies and cross allergies, indications and contraindications, gender medication review and repeated medication.
3 Approach
Figure 1 is an overview of our method, with four main stages: (i) data collection and pre-processing (Sect. 3.1), (ii) development of domain ontology (Sect. 3.2), (iii) knowledge extraction (Sect. 3.3), and (iv) data warehousing (Sect. 3.4).
In the first stage, the raw drug data is obtained through data crawler and preprocessing; In the second stage, the ontology of MKGis developed by manual induction from the raw drug data; In the third stage, the ontology is used for formulating entity and relation labeling scheme in the raw drug data so as to get the manual labeled dataset, and then the manual labeled dataset is used for the model training, rule designing and knowledge extracting; Finally, the extracted knowledge is stored in the KGafter disambiguation and format standardization.
3.1 Data Collecting and Preprocessing
Our data mainly come from three aspects: drug instructions, authoritative books and relevant medical literature.
The drug instruction is a document with legal effect, which contains all the information on how to use the drug safely and effectively. It is the most basic and important information for doctors’ prescriptions and patients’ safe medication [7], and it is also an important basis for RDU. However, the format of this kind of data is not unified and can only be used after structured processing. There are a large number of drug instructions on websites such as China listed drug catalogue [8] and China medical information query platform [9], etc. We use the scrapy framework to crawl the web pages that contain the drug instructions, then convert them into the form of key value pairs after filtering tags and special symbol, such as General name: Ganmao Qingre Granule, finally we remove duplication.
Authoritative books refer to the e-books containing pharmaceutical knowledge required for RDUand the existing medical concept classification system, such as The Foundation and Clinical Application of Drug Interaction [10]. The data form is standardized and rigorous. We compile matching rules according to the typesetting format for knowledge extraction.
Other literature is mainly about the research of a certain kind of drugs or the classification system of a certain medical concept. This kind of data has the characteristics of low density, different expressions and closer to natural language. It needs to be extracted manually and transformed into structured knowledge.
3.2 Development of Domain Ontology
The development methods of domain ontology are different, including Iterative Approach [11], ENTERPRISE Method [12], etc. In order to improve the effect of conceptual knowledge required for RDUiteratively and carry out verification rapidly, we attempt an Iterative Approach for developing medical ontology, as shown below:
-
1
Determine the domain of the ontology. Our ontology is used to model the RDUin the medical subdivision field, which is used to deal with the requirements involved in RDU.
-
2
Consider reusing existing ontologies. We investigate the existing MKG[1,2,3,4,5, 13,14,15,16], and reuse some of these concepts. For example, symptom, pharmaceutical ingredient, etc.
-
3
Enumerate important terms in the ontology. After collecting the literature such as The Foundation and Clinical Application of Drug Interaction [10], we list the important terms in ontology.
-
4
Define the classes and the class hierarchy. We use a combination of top-down and bottom-up development methods to obtain domain concepts. That is, firstly, through the conceptual terms obtained in Step 3, we select the important and core concepts (disease, drug, etc.), then summarize or deduce them and associate them with some other concepts.
-
5
Define the properties of classes (slots). For example, the population includes age, gender, weight, etc. The Step 4 and Step 5 are carried out simultaneously.
-
6
Define the facets of the slots. That is, an attribute can have constraints such as value type, value range and number of values. For example, gender only includes the two values: male and female.
-
7
Create instances. We evaluate whether the ontology is complete and meets the requirements by instantiating a class. If it needs to be improved, the above steps are repeated.
Specifically, the process is shown in Fig. 2.
The KGontology we design contains 22 concepts, 2 general relations (subclass of and instance of), and 29 domain relations (component, interaction, etc.). For the peculiar relation in the field, it focuses on the drug itself and the relation with disease, symptom and other concepts to support the functional needs of RDU. Part of ontology concepts and relations examples are shown in Fig. 3.
The rounded rectangle in Fig. 3 represents the concept, while the text on the corresponding line represents the relation between concepts. For some review functions of RDU, it is necessary to calculate and compare the quantitative entities such as usage and dosage. For calculation conveniently, we subdivide the concepts involving quantity (blue rounded rectangle), such as specification, age, weight, frequency, duration, dose and maximum dose, into numerical values and quantifiers, and establish a quantifier table and conversion method for unifying the unit of measurement. The concepts of drug interaction result, drug compatibility result, medication result and incidence (light orange rounded rectangle) are introduced to represent the irrational medication results given in the drug instructions and other reference literature, so as to quickly determine the review results.
The concept classification system (subclass of relation) is omitted in this figure. There are six concepts (dark orange rounded rectangle) with classification system, including drug, population, route of administration, disease, symptom and physiological test. The classification system is designed by referring to relevant literature and sorting out a large number of drug instructions from bottom to top.
Due to the complexity of medication for different patients and different situations, medical treatment can not be expressed by simple triples. In order to represent complex knowledge, we define composite relation types. Multiple relations are included under the compound relation. For example, the medication relation includes medication usage, medication frequency, etc. In Freebase, this kind of relation is called CVT (Compound Value Type) [17], and in Wikidata, this kind of relation is called qualifier [18].
3.3 Knowledge Extraction
Knowledge extraction is divided into two steps: named entity recognition and relation extraction.
3.3.1 Named Entity Recognition
Medical texts contain a large number of medical terms, one of the difficulties of entity recognition is the determination of entity boundary. Combining with lexicon information performs well in entity recognition tasks [19]. We implement a named entity recognition model based on this strategy.
We concatenate the hidden layer feature H of MC-BERT [20] and the lexicon feature W as the input N, then use the BiGRU-CRF to recognize named entities in medical text. The lexical embedding \(W_i\) of the ith character c in the medical text is calculated by Formula 1 and Formula 2:
where
[a; b] means concatenate vector a and b. Set D is the set of all lexicons that containing the ith medical text character c. These lexicons come from an additional word2vector dictionary. The values of D are \(B_i\) (take c as the begin character), \(M_i\) (take c as the middle character), \(E_i\) (take c as the end character) and \(S_i\) (c becomes a single lexicon) respectively. z(c) indicates the frequency of the c in the training set, and e(c) indicates the word2vector of the c, which dimension is 50.
Because of the similar linguistic characteristics in medical terms, it is effective for extracting medical entities by introducing word information of characters.
3.3.2 Relation Extraction
In data processing stage, we have divided the drug instructions into short sentences with different categories according to their directory structure. Therefore, when there is a pair of entities in the sentence, the relation can be identified easily through the category of the entity pair in the statement. Further more, The relative position of entity pair is also helpful to distinguish their relation. For example, in the sentence crude polysaccharide 240 mg, adenosine 235 mg. is content, the four entities crude polysaccharide, 240 mg, adenosine, and 235 mg can be formed to two pairs with the relation of content because of the entity category. Then, the two entities close to each other are classified into the same entity relation triple.
Based on the above reasons, we propose a relation extraction model MF-PCNN, which is based on Piece-Wise CNN (PCNN) and integrates entity category information and character position information (Mutil-Feature, MF). Similar to the named entity recognition model, its basic text context feature comes from the hidden layer output H of MC-BERT, and we concatenate the character relative position embedding P as the input R. P is a matrix with the dimension of \((2m+1)\times d_p\), which participates in training together with the model after random initialization, and we set \(d_p\) to 64. The position embedding of ith character in the medical text is the \(p_i\)th row in matrix P. m is the length of current medical text. The model input feature R is calculated by the Formula 4 and Formula 5:
\(i_e\) represents the position of entity e in the statement, when calculating the relative position with the subject. We finally get \(P_{sub}\), and the relative position with the object is \(P_{obj}\).
We use two CNNs to capture the preorder and postorder features of entities respectively. One of them is for processing the start position of statement to the position of the second entity, and the other is for processing the position of the first entity to the end position of statement. Then we max pooling them in the text direction. As shown in Formulas 6 and 7.
R[a : b] represents the segment from a to b in the feature R, \(i_{sub}\) is the position of subject in the text and \(i_{obj}\) is the object’s. Then we concatenate \(O_{pre}\), \(O_{post}\) and the entity category embedding E (Formula 8). Finally we use linear layer to reduce the dimension and use softmax() function to classify the relation, as shown in Formula 9.
\(E_{sub}\) and \(E_{obj}\) are the category embedding of subject and category embedding of object, respectively. The dimension of E is \(c_e\times d_e\). Its construction method is similar to that of P. \(c_e\) is the number of entity categories. W and B are the weight matrix and offset vector of linear layer, respectively. We choose cross entropy as the loss function to train the model.
3.4 Data Warehousing
3.4.1 Data Disambiguation
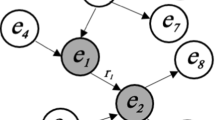
We choose the Neo4j as a graph database for data storage. In order to solve the ambiguity of storage and query between multi-hop relation, composite nodes are used. As shown in Fig. 4, since it is impossible to determine whether dose 1 or dose 2 is used by population 1, we choose composite nodes to solve this kind of problem.
We first define a set of multi-hop relation as a piece of knowledge. The first subject is called the head node, the last object of the chain is called the tail node, and other entities are collectively called the intermediate node. The head node is no longer connected with the intermediate node and the tail node, but directly connected with a composite node. Then, the composite node is connected with the intermediate node and tail node in this group of multi-hop relations. Because each tail node corresponds to a unique composite node, the drug dose used by different populations can be uniquely determined through the composite node.
3.4.2 Entity Standardization and Alignment
In order to reduce the workload of manual labeling, when it comes to entity labeling with numbers and units, we label values and quantifiers uniformly. However, in drug review, we need to use numerical values and quantifiers to review dosage, age and other information. Therefore, we refine some entities and extract the numerical values and quantifiers.
In such entities, we first convert Chinese numbers into Arabic numerals, and add the default number“1”next to some special characters, such as each, each time, etc. Then two types of half are processed: one and a half pieces is processed as 1.5 pieces and half pieces is processed as 0.5 pieces. Then we use regular expressions to match numerical values and quantifiers: match numerical values according to some connecting symbols such as“−”in \(2-3\) pieces, match quantifiers by using the position in the text, and finally convert units.
Because the medical text is rigorous and has rich domain characteristics, the expression of medical terms is usually unique. In the entity alignment, we use the string matching method to align the medical entities such as drug, disease, etc. The entities with semantic information such as population are classified by calculating the word vector of cosine similarity between the standard expression. For entities such as frequency, we use the above rule-based method to process them.
4 Experimental Setup
4.1 Dataset
Our purpose is to construct an MKGto meet the needs of RDU, which requires an amount raw medical data. In order that the knowledge extraction method can process these raw data, a labeled dataset should be built. Then we build a medical dataset by manual labelling.
We collect and preprocess three types of data including drug instructions, authoritative books and relevant medical literature. The number of semi-structured or structured data sorted out is 119,631 in the form of key: value. In the semi-structured data, valuable information can be obtained according to its directory name. For example, the component of a drug may contain drug, pharmaceutical ingredient or content. The steps of labeling this kind of data are as follows.
-
1
Define the entity and relation categories, which need to be labeled according to our domain ontology.
-
2
Unify the directory names and select the directories with high frequency, such as component, treatment, drug interaction, drug adverse reaction, specification, unsuitable population, contraindication, attention and medication, as the directories to be labeled.
-
3
Remove stop words and filter special characters.
-
4
Segment long sentences into short sentences if the sentence contains more than 128 characters. The number of short sentences to be labeled is 28,212.
-
5
Use brat [21] system to label manually.
-
6
Adopt cross validation to avoid mislabeling and wrong labeling, that is, after each annotator labels, another annotator checks the labeled content.
Figure 5 is an example of labeling a drug instruction document by brat.
There are 160,841 entities and 145,235 relations in labeled dataset. The statistics of entities or relations in different categories are shown in Figs. 6 and 7. There is a long tail phenomenon in entities and relations. Through the actual situation in the labeling process, we find that the main reason is that some entities or relation categories are indeed rare in the dataset. We divide the dataset into 80:10:10 for training, verification and testing.
4.2 Metrics
Assuming that \(S_t\) is the real label set and \(S^{'}_t\) is the label set predicted by the model, where t represents all data with category t. For the data of category t, \(P_t\) (Precision), \(R_t\) (Recall) and \(F1_t\) can be calculated from Formula 10, Formula 11 and Formula .
We use Micro-F1 and Macro-F1 as the overall evaluation indicators of the model, which are represented by Formulas 13 and 14 respectively.
where |T| represents the number of elements in T, P is calculated by Formula 15 and R is calculated by Formula 16.
5 Results and Insights
In our approach, the development of domain ontology is the step with methodology, which has been described in the Sec. 3.2, and the methods and examples of data warehousing are also described in Sec. 3.4.
All the experiments of knowledge extraction are implemented in the environment with 64G memory and Titan V GPU. To compare the effect of named entity recognition of each model, the P, R and F1 score are used as evaluation indicator. The input features of all models are from MC-BERT and all the results are after 50 rounds of training.
Overall results of named entity recognition are shown in Table 1, which are calculated by Formula 13 and Formula 14. It can be seen that the Lexicon-BiGRU-CRF obtains the optimal effect, which proves the effectiveness of BiGRU-CRF model with lexical information in medical text named entity recognition task. CRF [22] only uses the hidden layer feature of MC-BERT. Due to the lack of RNN structure to capture the context features of text, the overall effect of entity recognition is poor. Huang et al. [23] proposed BiLSTM-CRF for named entity recognition, which has achieved good results in recent years and is used as the baseline model of named entity recognition. We implement it and also use BiGRU to replace the BiLSTM structure. BiGRU-att-CRF [24] adds attention [25] (att) on the BiLSTM-CRF, but the result is not ideal because MC-BERT already has this kind of mechanism. We also explore different combinations (add, cat and pl) of the features between attention and BiGRU, where“add”means that these two types of features are added and input into CRF,“cat”means concatenating these two features, and“pl”means taking the output of BiGRU as the input of attention.
This further shows that the vocabulary or word information has a good improvement on data with strong domain characteristics such as medical corpora in named entity recognition. For example, entities of Pharmaceutical ingredient usually have a fixed way of expression, when the target entity contains a structure similar to lexical information. This method can more effectively identify the boundary of this entity. In order to further illustrate this problem, we use the models to carry out experiments on different entity categories. The main results are shown in Table 2, and the implementation evaluation indicator is shown in Formula 12.
As can be seen from Table 2, the model Lexicon-BiGRU-CRF performs well for most categories of entities. Among them, the instances of Incidence, Disease and Symptom (Fig. 3 shows that Disease and Symptom have the same status, so we treat them as a whole), Population and Pharmaceutical ingredient are usually standardized and limited, so they will get better results after using lexical information. For example, the two entities of Pharmaceutical ingredient: clindamycin and clindamycin phosphate. Both of them contain the word clindamycin. When the external lexicon contains such words, the model can more easily judge that the boundary of the two entities is clindamycin. Entity Dosage is expressed regularly in the sentence. For example, take two pills at a time, take three tablets a day, etc. BiGRU in the model can more effectively capture these expressions with similar structure, so as to improve the recognition accuracy of such entities. The recognition scores of all models for entity Etiology are not high, because it contains a variety of entities with practical significance, such as surgery, injury, microorganism and so on. Even so, our model still performs the best.
We use the model MF-PCNN for relation extraction and compare it with other baseline models. The experimental results are calculated by Formula 13 and Formula 14, which are shown in Table 3.
The RNN based model can extract the context information of text. We use BiGRU as the RNN structure and max pooling the out feature to build the first model in Table 3. BiLSTM-att [26] replaces the max pooling with attention, which calculates the attention weight of the BiLSTM output to obtain a feature vector of relation extraction. CNN model can capture local features of text. We also use max pooling technology and attention to implement two kinds of CNN based models. In order to make full use of the contextual and local features of text, RNN and CNN are usually used together. We take the output of BiGRU as the input of CNN, and then use max pooling to obtain the vector with context and local features, which is used as the feature vector of relationp category. PCNN [27] introduces the relative position information of the entity and the text features around the entity by encoding different segments of the text, which has achieved good results in relation extraction.
The above six models are used as baseline models to compare with our model MF-PCNN. In contrast, our model integrates more information, including the category of entities and the relative location of characters. The final results show that the model, MF-PCNN, performs well in the task of medical text relation extraction. In order to make a more detailed comparison, we do fine-grained extraction experiments on different relation categories. The main results are shown in Table 4, which are calculated from Formula 12.
In relation extraction of medical text, the relative position between characters is very important. For example, in the text very few cases have fainting, mostly due to too fast injection. and the text most cases have fainting, and very few are due to too fast injection. The two Incidence entities of very few and mostly/most only exchange the order, which eventually leads to a completely different meaning of the entity relation triplet between them and the Disease and Symptom entity fainting. When the relative position information is used, the extraction effect of our model in relation categories such as result is improved. In addition, the category of entities is also helpful for the determination of some relations. For example prescribe only represents the relation between entities Drug and Population, suffer from represents the relation between entities Population and Disease and Symptom, and so on. Some models in Table 4 got the same results. This is because this kind of relation is relatively simple. The extraction effect cannot be improved by increasing the complexity of the model. It is necessary to introduce external knowledge to improve the extraction effect of the model.
Since our raw data is semi-structured, it is unnecessary to list some entities and relations in Table 2 and Table 4. For example, for the text specification: 500ml, we can extract the Specification entity 500ml by the key-value. At the same time, when we know the category of some entities and the key of data, we can obtain their relation directly. For example, when the key of the data is“main component”, the Pharmaceutical ingredient entities obtained from the data have the component relation with the current Drug. The extracted data is stored after format standardization. Finally, we get 297,501 entities and 394,373 relations.
6 Related Work
6.1 Named Entity Recognition
At present, the mainstream entity recognition algorithm is a kind of deep learning method, which takes neural network as feature extractor and combines classifier to decode entity tags. Huang et al. [23] proposed a series of sequence labeling methods based on LSTM, including LSTM, BiLSTM, LSTM-CRF and BiLSTM-CRF. This is the first time that LSTM has been applied for sequence labeling in NLP. In the field of medical entity recognition, Ye et al. [28] integrated linguistic symbols, parts of speech (POS) and word structure features, and used conditional random field (CRF) to identify three types of named entities: diseases, clinical symptoms and surgical operations in 250 Chinese medical records. CRF was also adopted [29, 30] to extract the symptoms, pathogenesis and medical records in ancient medical records in the Ming and Qing Dynasties. Xue [31] introduced LSTM into the medical field, and combined transfer learning to improve the effect of the model through cross domain knowledge. Further more, the BiLSTM-CNN-CRF model [32] has achieved good recognition results on the Chinese electronic medical record (EMR) dataset of CCKS 2017.
6.2 Relation Extraction
In the early stage, researchers adopted machine learning methods and designed features manually to extract relations. Some methods [33,34,35] utilized different features such as the information of punctuation, word, context, entity and syntactic dependency to improve the extraction effect. In 2012, Richard et al. [36] proposed a deep learning model based on recurrent neural network (RNN) for relation extraction. After that, in 2014, a convolutional neural network (CNN) based model has been introduced into this field by Zeng et al. [37] for modeling local information of sentence. The subsequent works [38, 39] were also to modify the structure of the feature extractor based on the above two types of models.
6.3 Knowledge Graph
Initially, most of KGwere general KGthat constructed manually, such as WordNet [40] and opencyc [41]. The development of the Internet has promoted the emergence of new kinds of knowledge bases, which based on Internet resources, such as DBpedia [42], Yago [43], Freebase [17] and Wikidata [18].
Automated medical process supervision is based on MKG. Gong et al. [44] combined MKGwith EMR data to implement recommending medication. Compared with RDU, fewer factors are considered in medication recommendation. Relatively, the functions of RDUneed the support of relations between drugs, diseases, patients and treatment regimens, etc. At present, the researches of KGin Chinese medical field mainly includes [1,2,3,4,5, 7, 13,14,15,16, 45, 46]. Among them, researches on diseases [1,2,3,4,5, 13] are aimed at brain disease, diabetes, lung disease, heart disease, nephropathy and acne. Lin et al. [7] discussed the development method of ontology with the adverse reactions of quinolones. Nima et al. [45] implemented named entity recognition method on Tibetan medicine and constructed the KG. The prescription review [46] was based on the KGof Chinese patent medicine, which is only aimed at Chinese patent medicine and diseases, and did not implement the function of usage and dosage in RDU.
The construction methods of MKG[1,2,3,4,5, 13,14,15,16] usually study from a sub concept such as drug, symptom, disease and so on. In contrast, RDUdeals with the concepts of medical processes and their relations. On the other hand, RDUis of great significance in the process of clinical drug use, which can save medical resources, reduce adverse drug reactions caused by drug interactions, and play a strong guarantee for the safety of patients. Thus the construction of MKGis a subject that to be studied and developed urgently. Based on this, we studied the structure and expression of medical literature, drug instructions and other texts containing RDUinformation. Then by combination with the existing class hierarchy of medical concepts, we constructed an MKGfor RDU.
7 Discussion and Conclusion
Based on the texts containing medication information such as drug instructions, and combined with the needs of RDU, we designed the MKGontology for RDU. According to this ontology, we built a medical dataset for the training of knowledge extraction model. Then we implemented a named entity recognition model based on lexical information, and proposed a relation extraction model with multi feature fusion. After that, the entity relation triples were obtained. We disambiguated and aligned them, then stored them into the database and built the MKG.
Automated medical process supervision includes many subdivided fields such as RDU, medical insurance supervision and medical institutions supervision, etc. We have made a preliminary exploration in the field of RDU. The implementation of RDUmay be based on expert rules, recommendation, etc. Because this field contains a lot of medical concepts and knowledge, we choose KGas a specific tool for the review of prescription.
As an engineering problem, construction of KGis usually closer to practical application. Guided by the needs of indication examination, repeated drug use examination, review of drug interaction, adverse reaction query, review of precautions, usage and dosage review in the RDU, we constructed a MKG. Our KGfocuses on drugs and organizes concepts and relations with diseases and population as the main body. Therefore, this KGis more inclined to the auxiliary supervision of clinical medication.
The knowledge in the medical field is highly professional and has low tolerance for errors. Building a complete KGto meet all the needs of RDUcan not be achieved overnight. Our research still ignores some complex pharmaceutical knowledge, such as pharmacokinetics and drug toxicology. The medical text corresponding to these knowledge usually has more characteristics of natural language. If we want to use a knowledge representation framework to structure it losslessly, it is inseparable from the joint efforts of medical experts and computer experts. On the other hand, we simplified some complex relations between concepts in the ontology. For example, it may be necessary to determine the time of intravenous drip in the route of administration, but our ontology does not involve these kind of knowledge.
To sum up, the MKGwe constructed meets the basic needs of RDU, which can be used as a reference and promote the research in RDUfield. Meanwhile, developing a complete ontology of RDUis still the difficulty and the hot spot in the construction of MKG, which also requires a large number of scholars and even medical experts to study, and we will conduct further research based on this in the future.
Availability of Data and Materials
All resources from this project are available at https://github.com/zhenjia2017/RDUKG.
References
Hong L, Shi XY. A method for constructing medical ontology: the case of Brain Area and Autism. J Inf Resour Manag. 2020;10(2):80–90.
Liu H. X, Research on Construction and Retrieval of Diabetes Ontology, Master’s thesis, Jilin University, 2014.
Jia XH, Song WA, Li WY, Wang Q, Lei Y, Chen ZH, Chang ZP. Research and Implementation of the Construction of Slow Obstructive Pulmonary Knowledge Map. J Chin Comput Syst. 2020;41(7):1371–4.
Fu Y, Liu MF, Qiao R. Construction of Chinese Knowledge Graph of Heart Disease. J Wuhan Univ (Science Edition). 2020;66(3):261–7.
Lin YR, Zhang Y, Liu D, Qiao DP, Si HY, Jiang YP, Zhu J, Lu KD, Cheng H. Constructing a Medical Knowledge Graph of Nephropathy Based on the Electronic Medical Records of Nephropathy Specialists. J Southwest Univ (Natural Science Edition). 2020;42(11):52–8.
Standing Committee of the National People’s Congress. Pharmaceutical Administration Law of the People’s Republic of China. Gazette of the Standing Committee of the National People’s Congress of the People’s Republic of China. 2019;5(2):771–88.
Lin X, Li XY, Guo JJ, Zhang HB, Zhao JW, Ren HL. Investigation on the Construction of Domain Ontology in the Field of Adverse Drug Reaction of Quinolones. Digital Library Forum. 2020;191(4):39–46.
Yoozh Net, Catalogue of Listed Pharmaceuticals in China, website, 2021, https://db.yaozh.com/directory.
China Medical Information Platform, Authoritative Medical Information Query, website, 2021, https://www.dayi.org.cn.
Liu ZJ, Han HL. The Foundation and Clinical Application of Drug Interaction. 3rd ed. Beijing, China: People’s Medical Publishing House; 2019.
Natalya F. N, Deborah L. M, Ontology Development 101: A Guide to Creating Your First Ontology, Knowledge Systems Laboratory, vol. 32, p. 25, Jan 2001.
Li J, Meng LS. Comparison of Seven Approaches in Constructing Ontology. New Technol Lib Inf Serv. 2004;112(7):17–22.
Cui YD, Wang MQ, Chen XR, Zhang L, Li GZ. Construction for Ontology of Acne in Traditional Chinese Medicine, Modernization of Traditional Chinese Medicine and Materia Materia -. World Sci Technol. 2019;21(12):2867–72.
Lu K. Z, The Construction and Application of Knowledge Graph based on the Ancient Books of Traditional Chinese Medicine, Master’s thesis, Beijing Jiaotong University, 2020.
Wu H, Study on The Construction and Application of TCM Knowledge Graph about Compendium of Materia Medica, Master’s thesis, Zhengzhou University, 2020.
Weng H, Liu Z. Q, Yan S. X, Fan M. Y, Ou A. H, Chen D. C, Hao T. Y, A Framework for Automated Knowledge Graph Construction Towards Traditional Chinese Medicine, in Proceedings of Health Information Science, S. Siuly, Z. S. Huang, U. Aickelin, R. Zhou, H. Wang, Y. C. Zhang, and S. Klimenko, Ed.Cham: Springer International Publishing, 2017, pp. 170–181.
Bollacker K, Cook R, Tufts P, Freebase: A Shared Database of Structured General Human Knowledge, in Proceedings of the 22nd National Conference on Artificial Intelligence - Volume 2, ser. AAAI’07. Vancouver, British Columbia, Canada: AAAI Press, 2007, p. 1962–1963.
Denny V, Markus K. Wikidata: A Free Collaborative Knowledgebase. Communications of the ACM. Sep 2014;57(10):78–85.
Ma R. T, Peng M. L, Zhang Q, Wei Z. Y, Huang X. J, Simplify the Usage of Lexicon in Chinese NER, in Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics. Online: Association for Computational Linguistics, Jul 2020, pp. 5951–5960.
Zhang N. Y, Jia Q. H, Yin K. P, Dong L, Gao F, Hua N. W, Conceptualized Representation Learning for Chinese Biomedical Text Mining, arXiv preprint arXiv:2008.10813, 2020.
Pyysalo S, Stenetorp P, Topić G, Ohta T, Brat Rapid Annotation Tool, website, 2021, http://brat.nlplab.org/index.html.
John L, Andrew M, Fernando C. N. P, Conditional Random Fields: Probabilistic Models for Segmenting and Labeling Sequence Data, proceedings of icml, 2002.
Huang Z. H, Xu W, Yu K, Bidirectional LSTM-CRF Models for Sequence Tagging, arXiv preprint arXiv: 1508.01991, 2015.
X. J. Xie, Z. Xie, K. Ma, J. G. Chen, Q. J. Qiu, H. Li, S. Y. Pan, and L. F. Tao, Geological Named Entity Recognition based on BERT and BiGRU-Attention-CRF Model, Geological Bulletin of China, pp. 1–13, 2021.
V. Ashish, S. Noam, P. Niki, U. Jakob, J. Llion, G. Aidan N., K. Łukasz, and P. Illia, Attention is All You Need, in Proceedings of the 31st International Conference on Neural Information Processing Systems, ser. NIPS’17. Red Hook, NY, USA: Curran Associates Inc., 2017, p. 6000–6010.
P. Zhou, W. Shi, J. Tian, and Z. Y. Qi, Attention-Based Bidirectional Long Short-Term Memory Networks for Relation Classification, in Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (Volume 2: Short Papers), 2016.
D. J. Zeng, K. Liu, Y. B. Chen, and J. Zhao, Distant Supervision for Relation Extraction via Piecewise Convolutional Neural Networks, in Conference on Empirical Methods in Natural Language Processing, 2015.
Ye F, Chen YY, Zhou GG, Li HM, Li Y. Intelligent Recognition of Named Entities in Electronic Medical Records. Chin J Biomed Eng. 2011;30(2):256–62.
Wang SK, Li SZ, Chen TS. Recognition of Chinese Medicine Named Entity Based on Conditional Random Field. J Xiamen Univ (Natural Science). 2009;048(3):359–64.
Li W, Zhao DZ, Li B, Peng XM, Liu JR. Combining CRF and rule based medical named entity recognition. Appl Res Comput. 2015;32(4):1082–6.
Xue T. Z, Research on Chinese Named Entity Recognition in Medical Field, Master’s thesis, Harbin Institute of Technology, 2017.
Liu YP, Li DD. Chinese Named Entity Recognition Method Based on Bi-directional LSTM-CNN-CRF. J Harbin Univ Sci Technol. 2020;25(1):115–20.
Sun X, Dong L. H, Feature-Based Approach to Chinese Term Relation Extraction, in Proceedings of 2009 International Conference on Signal Processing Systems (ICSPS). Los Alamitos, CA, USA: IEEE Computer Society, May 2009, pp. 410–414.
Che WX, Liu T, Li S. Automatic entity relation extraction. J Chin Inf Process. 2005;19(2):1–6.
Gan LX, Wan CX, Liu DX, Zhong Q, Jiang TJ. Chinese entity and relation extraction based on syntactic and semantic analysis. J Comput Res Dev. 2016;53(2):284–302.
S. Richard, H. Brody, M. Christopher D., and N. Andrew Y., Semantic Compositionality through Recursive Matrix-Vector Spaces, in Proceedings of the 2012 Joint Conference on Empirical Methods in Natural Language Processing and Computational Natural Language Learning, ser. EMNLP-CoNLL’12. USA: Association for Computational Linguistics, 2012, p. 1201–1211.
D. J. Zeng, K. Liu, S. W. Lai, and G. Y. Zhou, and J. Zhao, Relation Classification via Convolutional Deep Neural Network, in Proceedings of COLING 2014, the 25th International Conference on Computational Linguistics: Technical Papers. Dublin, Ireland: Dublin City University and Association for Computational Linguistics, Aug 2014, pp. 2335–2344.
Gao D, Peng DL, Liu C. Entity relation extraction based on CNN in large-scale text data. J Chin Comput Syst. 2018;39(5):1021–6.
Y. Xu, L. L. Mou, G. Li, Y. C. Chen, H. Peng, and Z. Jin, Classifying Relations via Long Short Term Memory Networks along Shortest Dependency Paths, in Proceedings of the 2015 Conference on Empirical Methods in Natural Language Processing.Lisbon, Portugal: Association for Computational Linguistics, Sep 2015, pp. 1785–1794.
George A. M, WordNet: A Lexical Database for English, Communications of the ACM, vol. 38, no. 11, p. 39–41, Nov 1995.
Jordi C, Veda C. S, Vijayan S, Usability of upper level ontologies: The case of ResearchCyc, Data & Knowledge Engineering, vol. 69, no. 4, pp. 343–356, 2010.
L. Jens, I. Robert, J. Max, J. Anja, K. Dimitris, M. Pablo N., H. Sebastian, M. Mohamed, K. Patrick Van, and B. Christian, DBpedia - A Large-scale, Multilingual Knowledge Base Extracted from Wikipedia, Semantic Web, vol. 6, no. 2, pp. 167–195, 2015.
T. Rebele, F. Suchanek, J. Hoffart, J. Biega, E. Kuzey, and G. Weikum, YAGO: A Multilingual Knowledge Base from Wikipedia, Wordnet, and Geonames, in Proceedings of the Semantic Web – ISWC 2016, P. Groth, E. Simperl, A. Gray, M. Sabou, M. Krötzsch, F. Lecue, F. Flöck, and Y. Gil, Ed. Cham: Springer International Publishing, 2016, pp. 177–185.
Gong F, Wang M, Wang HF, Wang S, Liu MY. SMR: Medical Knowledge Graph Embedding for Safe Medicine Recommendation. Big Data Research. 2021;23: 100174.
Luosanggadeng, Z. Nima, D. Renzeng, and J. Suonan, Research on Tibetan Medicine Entity Recognition and Knowledge Graph Construction, in Proceedings of the 10th International Conference on Computer Engineering and Networks, Q. Liu, X. D. Liu, T. Shen, and X. S. Qiu, Ed. Singapore: Springer Singapore, 2021, pp. 1718–1724.
Xiong WP, Cao J, Zhou X, Du JQ, Nie B, Zeng ZJ, Li TC. Design and Evaluation of a Prescription Drug Monitoring Program for Chinese Patent Medicine based on Knowledge Graph. Evid-Based Complement Alternat Med. 2021;2021:1–8.
Funding
This work was supported by the National Key R &D Program of China (2020AAA0105101); Sichuan Key R &D project (2020YFG0035; 2021YFS0014; 2022YFH0020).
Author information
Authors and Affiliations
Contributions
Xiong Liao contributed in writing, ontology developing and the design and methodology of the study. Meng Liao contributed in entity processing. Andi Guo contributed in data disambiguating. Xinran Luo and Ziwei Li contributed in data collecting and preprocessing. Other authors assisted with the production of the manuscript’s draft version. All authors examined the results and gave final approval to the manuscript’s final version.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare they have no conflicts of interest.
Ethical Approval and Consent to participate
This article does not contain any studies with animals performed by any of the authors.
Consent for publication
The authors hereby consent to publication of the work
Authors’ information
Xiong Liao, Meng Liao, Andi Guo, Xinran Luo, Ziwei Li, Tianrui Li, Shengdong Du, and Zhen Jia are with the School of Computing and Artificial Intelligence, Southwest Jiaotong University, Chengdu, 610031, China. Weiyuan Chen is with Sichuan Yice Science and Technology Co., Ltd, Chengdu, 610041, China.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liao, X., Liao, M., Guo, A. et al. Medical Knowledge Graph to Promote Rational Drug Use: Model Development and Performance Evaluation. Hum-Cent Intell Syst 2, 1–13 (2022). https://doi.org/10.1007/s44230-022-00005-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s44230-022-00005-z