Abstract
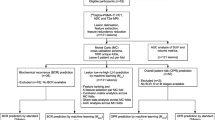
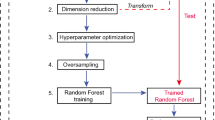
This study aimed to investigate the performance of 18F-DCFPyL positron emission tomography/computerized tomography (PET/CT) models for predicting benign-vs-malignancy, high pathological grade (Gleason score > 7), and clinical D'Amico classification with machine learning. The study included 138 patients with treatment-naïve prostate cancer presenting positive 18F-DCFPyL scans. The primary lesions were delineated on PET images, followed by the extraction of tumor-to-background-based general and higher-order textural features by applying five different binning approaches. Three layer-machine learning approaches were used to identify relevant in vivo features and patient characteristics and their relative weights for predicting high-risk malignant disease. The weighted features were integrated and implemented to establish individual predictive models for malignancy (Mm), high path-risk lesions (by Gleason score) (Mgs), and high clinical risk disease (by amico) (Mamico). The established models were validated in a Monte Carlo cross-validation scheme. In patients with all primary prostate cancer, the highest areas under the curve for our models were calculated. The performance of established models as revealed by the Monte Carlo cross-validation presenting as the area under the receiver operator characteristic curve (AUC): 0.97 for Mm, AUC: 0.73 for Mgs, AUC: 0.82 for Mamico. Our study demonstrated the clinical potential of 18F-DCFPyL PET/CT radiomics in distinguishing malignant from benign prostate tumors, and high-risk tumors, without biopsy sampling. And in vivo 18F-DCFPyL PET/CT can be considered a noninvasive tool for virtual biopsy for personalized treatment management.






Similar content being viewed by others
Abbreviations
- PET:
-
Positron emission tomography
- PCa:
-
Prostate cancer
- GS:
-
Gleason score
- PSA:
-
Prostate-specific antigen
- CT:
-
Computed tomography
- PSMA:
-
Prostate-specific membrane antigen
- TNM:
-
Tumor node metastasis
- MRI:
-
Magnetic resonance imaging
- CDSS:
-
Clinical decision support system
- SUV:
-
Standardized uptake value
- SUVmean:
-
Standardized uptake mean value
- SUVmax:
-
Standardized uptake max value
- SUVpeak:
-
Standardized uptake peak value
- SUVR:
-
Ratio of standardized uptake max value
- VOIs:
-
Volumes of interest
- PSMA-TV:
-
Total volume of PSMA
- TL-PSMA:
-
Total lesion of PSMA
- SMOTE:
-
Synthetic minority over-sampling technique
- IBSI:
-
Imaging biomarker standardization initiatives
- MC:
-
Monte Carlo
- ML:
-
Machine learning
- GLCM:
-
Gray-level cooccurrence matrix
- GLDZM:
-
Gray-level distance zone matrix
- NGLDM:
-
Neighboring grey level dependence matrix
- NGTDM:
-
Neighborhood grey tone difference matrix
- GLRLM:
-
Gray-level run-length matrix
- BYS:
-
Bayesian classification
- MGWC:
-
Multi-Gaussian weighting
- RF:
-
Random forest
- SVM:
-
Support vector machine
- WHO:
-
World Health Organization
- ISUP:
-
International Society of Urological Pathology
- UICC:
-
Union for International Cancer Control
- ACC:
-
Accuracy
- SENS:
-
Sensitivity
- SPEC:
-
Specificity
- PPV:
-
Positive predictive value
- NPV:
-
Negative predictive value
- AUC:
-
Area under the receiver operating characteristic curve
- DREs:
-
Digital rectal examinations
- M m :
-
Predictive models for malignancy
- M gs :
-
Predictive models for high path-risk lesions (by Gleason score)
- M amico :
-
Predictive models for high clinical risk disease (by amico)
- ROI:
-
Region of interest
References
Aerts HJ (2016) The potential of radiomic-based phenotyping in precision medicine: a review. JAMA Oncol 2(12):1636–1642. https://doi.org/10.1001/jamaoncol.2016.2631
Borofsky S, George AK, Gaur S, Bernardo M, Greer MD, Mertan FV, Taffel M, Moreno V, Merino MJ, Wood BJ, Pinto PA, Choyke PL, Turkbey B (2018) What are we missing? False-negative cancers at multiparametric MR imaging of the prostate. Radiology 286(1):186–195. https://doi.org/10.1148/radiol.2017152877
Bratan F, Niaf E, Melodelima C, Chesnais AL, Souchon R, Mege-Lechevallier F, Colombel M, Rouviere O (2013) Influence of imaging and histological factors on prostate cancer detection and localisation on multiparametric MRI: a prospective study. Eur Radiol 23(7):2019–2029. https://doi.org/10.1007/s00330-013-2795-0
Chen B, Zhang R, Gan Y, Yang L, Li W (2017a) Development and clinical application of radiomics in lung cancer. Radiat Oncol 12(1):154. https://doi.org/10.1186/s13014-017-0885-x
Chen S, Harmon S, Perk T, Li X, Chen M, Li Y, Jeraj R (2017b) Diagnostic classification of solitary pulmonary nodules using dual time (18)F-FDG PET/CT image texture features in granuloma-endemic regions. Sci Rep 7(1):9370. https://doi.org/10.1038/s41598-017-08764-7
D’Amico AV, Whittington R, Malkowicz SB, Schultz D, Blank K, Broderick GA, Tomaszewski JE, Renshaw AA, Kaplan I, Beard CJ, Wein A (1998) Biochemical outcome after radical prostatectomy, external beam radiation therapy, or interstitial radiation therapy for clinically localized prostate cancer. JAMA 280(11):969–974. https://doi.org/10.1001/jama.280.11.969
Gatenby RA, Grove O, Gillies RJ (2013) Quantitative imaging in cancer evolution and ecology. Radiology 269(1):8–15. https://doi.org/10.1148/radiol.13122697
Ghezzo S, Bezzi C, Presotto L, Mapelli P, Bettinardi V, Savi A, Neri I, Preza E, Samanes Gajate AM, De Cobelli F, Scifo P, Picchio M (2022) State of the art of radiomic analysis in the clinical management of prostate cancer: a systematic review. Crit Rev Oncol Hematol 169:103544. https://doi.org/10.1016/j.critrevonc.2021.103544
Hatt M, Tixier F, Pierce L, Kinahan PE, Le Rest CC, Visvikis D (2017) Characterization of PET/CT images using texture analysis: the past, the present... any future? Eur J Nucl Med Mol Imaging 44(1):151–165. https://doi.org/10.1007/s00259-016-3427-0
Hsieh PF, Chang TY, Lin WC, Chang H, Chang CH, Huang CP, Yang CR, Chen WC, Chang YH, Wang YD, Huang WC, Wu HC (2022) A comparative study of transperineal software-assisted magnetic resonance/ultrasound fusion biopsy and transrectal cognitive fusion biopsy of the prostate. BMC Urol 22(1):72. https://doi.org/10.1186/s12894-022-01011-w
Kyriakopoulos CE, Chen YH, Carducci MA, Liu G, Jarrard DF, Hahn NM, Shevrin DH, Dreicer R, Hussain M, Eisenberger M, Kohli M, Plimack ER, Vogelzang NJ, Picus J, Cooney MM, Garcia JA, DiPaola RS, Sweeney CJ (2018) Chemohormonal therapy in metastatic hormone-sensitive prostate cancer: long-term survival analysis of the randomized phase III E3805 CHAARTED Trial. J Clin Oncol 36(11):1080–1087. https://doi.org/10.1200/JCO.2017.75.3657
Ladjevardi S, Berglund A, Varenhorst E, Bratt O, Widmark A, Sandblom G (2013) Treatment with curative intent and survival in men with high-risk prostate cancer. A population-based study of 11,380 men with serum PSA level 20–100 ng/mL. BJU Int 111(3):381–388. https://doi.org/10.1111/j.1464-410X.2012.11320.x
Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, Sanduleanu S, Larue R, Even AJG, Jochems A, van Wijk Y, Woodruff H, van Soest J, Lustberg T, Roelofs E, van Elmpt W, Dekker A, Mottaghy FM, Wildberger JE, Walsh S (2017) Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 14(12):749–762. https://doi.org/10.1038/nrclinonc.2017.141
Laudicella R, Comelli A, Liberini V, Vento A, Stefano A, Spataro A, Croce L, Baldari S, Bambaci M, Deandreis D, Arico D, Ippolito M, Gaeta M, Alongi P, Minutoli F, Burger IA, Baldari S (2022) [(68)Ga]DOTATOC PET/CT radiomics to predict the response in GEP-NETs undergoing [(177)Lu]DOTATOC PRRT: the “Theragnomics” concept. Cancers (Basel) 14(4):984. https://doi.org/10.3390/cancers14040984
Le JD, Tan N, Shkolyar E, Lu DY, Kwan L, Marks LS, Huang J, Margolis DJ, Raman SS, Reiter RE (2015) Multifocality and prostate cancer detection by multiparametric magnetic resonance imaging: correlation with whole-mount histopathology. Eur Urol 67(3):569–576. https://doi.org/10.1016/j.eururo.2014.08.079
Litwin MS, Tan HJ (2017) The diagnosis and treatment of prostate cancer: a review. JAMA 317(24):2532–2542. https://doi.org/10.1001/jama.2017.7248
McGuire S (2016) World Cancer Report 2014. Geneva, Switzerland: World Health Organization, International Agency for Research on Cancer, WHO Press, 2015. Adv Nutr 7(2):418–419. https://doi.org/10.3945/an.116.012211
Nioche C, Orlhac F, Boughdad S, Reuze S, Goya-Outi J, Robert C, Pellot-Barakat C, Soussan M, Frouin F, Buvat I (2018) LIFEx: A freeware for radiomic feature calculation in multimodality imaging to accelerate advances in the characterization of tumor heterogeneity. Cancer Res 78(16):4786–4789. https://doi.org/10.1158/0008-5472.CAN-18-0125
Obek C, Doganca T, Demirci E, Ocak M, Kural AR, Yildirim A, Yucetas U, Demirdag C, Erdogan SM, Kabasakal L, Members of Urooncology Association T (2017) The accuracy of (68)Ga-PSMA PET/CT in primary lymph node staging in high-risk prostate cancer. Eur J Nucl Med Mol Imaging 44(11):1806-1812. https://doi.org/10.1007/s00259-017-3752-y
Papp L, Spielvogel CP, Grubmuller B, Grahovac M, Krajnc D, Ecsedi B, Sareshgi RAM, Mohamad D, Hamboeck M, Rausch I, Mitterhauser M, Wadsak W, Haug AR, Kenner L, Mazal P, Susani M, Hartenbach S, Baltzer P, Helbich TH, Kramer G, Shariat SF, Beyer T, Hartenbach M, Hacker M (2021) Supervised machine learning enables non-invasive lesion characterization in primary prostate cancer with [(68)Ga]Ga-PSMA-11 PET/MRI. Eur J Nucl Med Mol Imaging 48(6):1795–1805. https://doi.org/10.1007/s00259-020-05140-y
Perandini S, Soardi GA, Motton M, Augelli R, Dallaserra C, Puntel G, Rossi A, Sala G, Signorini M, Spezia L, Zamboni F, Montemezzi S (2016) Enhanced characterization of solid solitary pulmonary nodules with Bayesian analysis-based computer-aided diagnosis. World J Radiol 8(8):729–734. https://doi.org/10.4329/wjr.v8.i8.729
Perera M, Papa N, Christidis D, Wetherell D, Hofman MS, Murphy DG, Bolton D, Lawrentschuk N (2016) Sensitivity, specificity, and predictors of positive (68)Ga-prostate-specific Membrane antigen positron emission tomography in advanced prostate cancer: a systematic review and meta-analysis. Eur Urol 70(6):926–937. https://doi.org/10.1016/j.eururo.2016.06.021
Preisser F, Cooperberg MR, Crook J, Feng F, Graefen M, Karakiewicz PI, Klotz L, Montironi R, Nguyen PL, D’Amico AV (2020) Intermediate-risk prostate cancer: stratification and management. Eur Urol Oncol 3(3):270–280. https://doi.org/10.1016/j.euo.2020.03.002
Sarikaya S, Resorlu M, Oguz U, Yordam M, Bozkurt OF, Unsal A (2014) Evaluation of the pathologic results of prostate biopsies in terms of age, Gleason score and PSA level: our experience and review of the literature. Arch Ital Urol Androl 86(4):288–290. https://doi.org/10.4081/aiua.2014.4.288
Tan WP, Kotamarti S, Chen E, Mahle R, Arcot R, Chang A, Ayala A, Michael Z, Seguier D, Polascik TJ (2022) Oncological and functional outcomes of men undergoing primary whole gland cryoablation of the prostate: a 20-year experience. Cancer 128(21):3824–3830. https://doi.org/10.1002/cncr.34458
Valdez-Vargas AD, Sanchez-Lopez HM, Badillo-Santoyo MA, Maldonado-Valadez RE, Manzo-Perez BO, Perez-Abarca VM, Manzo-Perez G, Vanzzini-Guerrero MA, Alvarez-Canales JA (2021) Recurrence rate of localized prostate cancer after radical prostatectomy according to D'amico risk classification in a tertiary referral hospital: association study. Cir Cir 89(4):520–527. https://doi.org/10.24875/CIRU.200007601
Wang S, Dai Y, Shen J, Xuan J (2021) Research on expansion and classification of imbalanced data based on SMOTE algorithm. Sci Rep 11(1):24039. https://doi.org/10.1038/s41598-021-03430-5
Acknowledgements
The authors would like to thank Qilu Hospital of China, Vienna General Hospital of Austria, and Evomics Medical Technology Co., Ltd. for the support.
Author information
Authors and Affiliations
Contributions
YKL performed data curation, image analysis, investigation, methodology, original and revised draft writing and uploading; SLH and SWW performed machine learning analysis and original draft writing; FL and JN performed original draft writing; PS, JFL and SH performed radioactive tracer synthesis and quality control; LLQ performed image analysis; XL and XL performed methodology, writing, reviewing and editing.
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no competing interests to declare that are relevant to the content of this article.
Ethics approval
This study was approved by the Ethical Committee of Shandong University Qilu Hospital, Jinan, Shandong, China (KYLL-2017–573). All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Data availability
The datasets generated during analysis in the current study are available from the corresponding author.
Consent to participate
Written informed consent was obtained from each participant before enrollment.
Consent for publication
The participants have consented to the submission of the data and figures to the journal.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, Y., Li, F., Han, S. et al. Performance of 18F-DCFPyL PET/CT in Primary Prostate Cancer Diagnosis, Gleason Grading and D'Amico Classification: A Radiomics-Based Study. Phenomics 3, 576–585 (2023). https://doi.org/10.1007/s43657-023-00108-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43657-023-00108-y