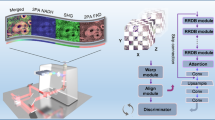
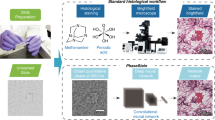
Abstract
Fluorescence labeling and imaging provide an opportunity to observe the structure of biological tissues, playing a crucial role in the field of histopathology. However, when labeling and imaging biological tissues, there are still some challenges, e.g., time-consuming tissue preparation steps, expensive reagents, and signal bias due to photobleaching. To overcome these limitations, we present a deep-learning-based method for fluorescence translation of tissue sections, which is achieved by conditional generative adversarial network (cGAN). Experimental results from mouse kidney tissues demonstrate that the proposed method can predict the other types of fluorescence images from one raw fluorescence image, and implement the virtual multi-label fluorescent staining by merging the generated different fluorescence images as well. Moreover, this proposed method can also effectively reduce the time-consuming and laborious preparation in imaging processes, and further saves the cost and time.








Similar content being viewed by others
Data Availability
Data underlying the results presented in this paper are not publicly available at this time but may be obtained from the authors upon reasonable request. The code is available within the supplementary information.
Abbreviations
- CNN:
-
Convolutional neural network
- DL:
-
Deep learning
- GAN:
-
Generative adversarial network
- H&E:
-
Hematoxylin and Eosin
- DAPI:
-
4′,6-Diamidino-2-phenylindole
- WGA:
-
Wheat germ agglutinin
- cGAN:
-
Conditional generative adversarial network
- MS-SSIM:
-
Multi-scale structural similarity
- CLAHE:
-
Contrast limited adaptive histogram equalization
- SSIM:
-
Structural similarity
- PSNR:
-
Peak signal-to-noise ratio
- MAE:
-
Mean absolute error
- NA:
-
Numerical aperture
- CMOS:
-
Complementary metal oxide semiconductor
References
Chan LL, McCulley KJ, Kessel SL (2017) Assessment of cell viability with single-, dual-, and multi-staining methods using image cytometry. Methods Mol Biol 1601:27–41. https://doi.org/10.1007/978-1-4939-6960-9_3
Cheng S, Fu S, Kim YM, Li Y, Xue Y, Yi J, Tian L (2021) Single-cell cytometry via multiplexed fluorescence prediction by label-free reflectance microscopy. Sci Adv 7:eabe0431. https://doi.org/10.1126/sciadv.abe0431
Christiansen EM, Yang SJ, Ando DM (2018) In silico labeling: predicting fluorescent labels in unlabeled images. Cell 197:792–803. https://doi.org/10.1016/j.cell.2018.03.040
Gatys LA, Ecker AS, Bethge M (2016) Image style transfer using convolutional neural networks. In: Conference on computer vision and pattern recognition (CVPR), pp 2414–2423. https://doi.org/10.1109/CVPR.2016.265
Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, Bengio Y (2014) Generative adversarial networks. Statistics. https://doi.org/10.48550/arXiv.1406.2661
Haan K, Zhang Y, Zuckerman J, Liu T, Sisk AE, Diaz MFP, Jen K, Nobori A, Liou S, Zhang S, Riahi R, Rivenson Y, Wallace WD, Ozcan A (2021) Deep learning-based transformation of H&E stained tissues into special stains. Nat Commun 12:4884. https://doi.org/10.1038/s41467-021-25221-2
Hertzmann A, Jacobs CE, Oliver N, Curless B, Salesin DH (2001) Image analogies. In: Conference on computer graphics and interactive techniques, pp 327–340. https://doi.org/10.1145/383259.383295
Horssen P, Siebes M, Hoefer I, Spaan J, Wijngaard J (2010) Improved detection of fluorescently labeled microspheres and vessel architecture with an imaging cryomicrotome. Med Biol Eng Comput 48:735–744. https://doi.org/10.1007/s11517-010-0652-8
Huang X, Liu MY, Belongie S (2018) Multimodal unsupervised image-to-image translation. In: European conference on computer vision (ECCV), pp 172–189. https://doi.org/10.1007/978-3-030-01219-9_11
Huang J, Liao J, Kwong S (2022) Unsupervised image-to-image translation via pre-trained styleGAN2. IEEE Trans Multimedia 24:1435–1448. https://doi.org/10.1109/TMM.2021.3065230
Imboden S, Liu X, Lee BS, Payne MC, Hsieh C, Lin NYC (2021) Investigating heterogeneities of live mesenchymal stromal cells using AI-based label-free imaging. Sci Rep 11:6728. https://doi.org/10.1038/s41598-021-85905-z
Isola P, Zhu J, Zhou T, Efros A (2017) Image-to-image translation with conditional adversarial networks. In: Conference on computer vision and pattern recognition (CVPR), pp 5967–5976. https://doi.org/10.1109/CVPR.2017.632
Jiang Z, Li B, Tran T, Jiang J, Liu X, Ta D (2022) Fluo-Fluo translation based on deep learning. Chi Opt Lett 20:031701. https://doi.org/10.3788/COL202220.031701
Klages P, Benslimane I, Riyahi S, Jiang J, Hunt M, Deasy JO, Veeraraghavan H, Tyagi N (2020) Patch-based generative adversarial neural network models for head and neck MR-only planning. Med Phys 47:626–642. https://doi.org/10.1002/mp.13927
LaChance J, Cohen DJ (2020) Practical fluorescence reconstruction microscopy for large samples and low-magnification imaging. Plos Comput Biol 16:e1008443. https://doi.org/10.1371/journal.pcbi.1008443
Lahiani A, Navab N, Albarqouni S, Klaiman E (2019) Perceptual embedding consistency for seamless reconstruction of tilewise style transfer. In: Medical image computing and computer assisted intervention (MICCAI), pp 568–576. https://doi.org/10.1007/978-3-030-32239-7_63
Lahiani A, Klaman I, Navab N, Albarqouni S, Klaiman E (2021) Seamless virtual whole slide image synthesis and validation using perceptual embedding consistency. IEEE J Biomed Health 25:403–411. https://doi.org/10.1109/JBHI.2020.2975151
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521:436–444. https://doi.org/10.1038/nature14539
Li D, Hui H, Zhang Y, Tong W, Tian F, Yang X, Liu J, Chen Y, Tian J (2020) Deep learning for virtual histological staining of bright-field microscopic images of unlabeled carotid artery tissue. Mol Imaging Biol 22:1301–1309. https://doi.org/10.1007/s11307-020-01508-6
Li X, Zhang G, Qiao H, Bao F, Deng Y, Wu J, He Y, Yun J, Lin X, Xie H, Wang H, Dai Q (2021) Unsupervised content-preserving transformation for optical microscopy. Light Sci Appl 10:44. https://doi.org/10.1038/s41377-021-00484-y
Liu M, Huang X, Mallya A, Karras T, Aila T, Lehtinen J, Kautz J (2019) Few-Shot unsupervised image-to-image translation. In: International conference on computer vision (ICCV), pp 10550–10559. https://doi.org/10.1109/ICCV.2019.01065
Mao Q, Lee H, Tseng H, Ma S, Yang M (2019) Mode seeking generative adversarial networks for diverse image synthesis. In: Conference on computer vision and pattern recognition (CVPR), pp 1429–1437. https://doi.org/10.1109/CVPR.2019.00152
McCann MT, Jin KH, Unser M (2017) Convolutional neural networks for inverse problems in imaging: a review. IEEE Signal Process Mag 34(85–95):2017. https://doi.org/10.1109/MSP.2017.2739299
Mirza M, Osindero S (2014) Conditional generative adversarial nets. Comput Sci. https://doi.org/10.48550/arXiv.1411.1784
Muniyappan S, Allirani A, Saraswathi S (2013) A novel approach for image enhancement by using contrast limited adaptive histogram equalization method. In International conference on computing, communications and networking technologies (ICCCNT), pp 1–6. https://doi.org/10.1109/ICCCNT.2013.6726470
Reza AM (2004) Realization of the contrast limited adaptive histogram equalization (CLAHE) for real-time image enhancement. J Vlsi Sig Proc Syst 38:35–44. https://doi.org/10.1023/B:VLSI.0000028532.53893.82
Rivenson Y, Liu T, Wei Z, Zhang Y, Haan K, Ozcan A (2019a) PhaseStain: the digital staining of label-free quantitative phase microscopy images using deep learning. Light Sci Appl 8:23. https://doi.org/10.1038/s41377-019-0129-y
Rivenson Y, Wang H, Wei Z, Haan K, Zhang Y, Wu Y, Gunaydin H, Zuckerman JE, Chong T, Sisk AE, Westbrook LM, Wallace WD, Ozcan A (2019b) Virtual histological staining of unlabelled tissue-autofluorescence images via deep learning. Nat Biomed Eng 3:466–477. https://doi.org/10.1038/s41551-019-0362-y
Rivenson Y, Haan K, Wallace WD, Ozcan A (2020) Emerging advances to transform histopathology using virtual staining. BME Frontiers 2020:9647163. https://doi.org/10.34133/2020/9647163
Ronneberger O, Fischer P, Brox T (2015) U-Net: convolutional networks for biomedical image segmentation. In: Medical image computing and computer-assisted intervention (MICCAI), pp 234–241. https://doi.org/10.1007/978-3-319-24574-4_28
Rouse DM and Hemami SS (2008) Analyzing the role of visual structure in the recognition of natural image content with multi-scale SSIM. In: Proc. SPIE, vol 6806, pp 680615. https://doi.org/10.1117/12.768060
Seldenrijk CA, Morson BC, Meuwissen SG, Schipper NW, Lindeman J, Meijer CJ (1991) Histopathological evaluation of colonic mucosal biopsy specimens in chronic inflammatory bowel disease: diagnostic implications. Gut 32:1514–1520. https://doi.org/10.1136/gut.32.12.1514
Shigene K, Hiasa Y, Otake Y, Soufi M, Janewanthanakul S, Nishimura T, Sato Y, Suetsugu S (2021) Translation of cellular protein localization using convolutional networks. Front Cell Dev Biolfront 9:635231. https://doi.org/10.3389/fcell.2021.635231
Wang Z, Bovik AC, Sheikh HR, Simoncelli EP (2004) Image quality assessment: From error visibility to structural similarity. IEEE Trans Image Process 13:600–612. https://doi.org/10.1109/tip.2003.819861
Zhang R, Isola P, Efros AA (2016) Colorful image colorization. In: European conference on computer vision (ECCV), pp 649–666. https://doi.org/10.1007/978-3-319-46487-9_40
Zhang Y, Li K, Li K, Wang L, Zhong B, Fu Y (2018) Image super-resolution using very deep residual channel attention networks. In: European conference on computer vision (ECCV), pp294–310. https://doi.org/10.1007/978-3-030-01234-2_18
Zhang Y, De H, Rivenson Y, Li J, Delis A, Ozcan A (2020) Digital synthesis of histological stains using micro-structured and multiplexed virtual staining of label-free tissue. Light Sci Appl 9:78. https://doi.org/10.1038/s41377-020-0315-y
Zhao H, Gallo O, Frosio I, Kautz J (2017) Loss functions for image restoration with neural networks. IEEE Trans Comput Imag 3:47–57. https://doi.org/10.1109/TCI.2016.2644865
Zhu J, Park T, Isola P, Efros AA (2017a) Unpaired image-to-image translation using cycle-consistent adversarial networks. In: IEEE international conference on computer vision (ICCV), pp 2242–2251. https://doi.org/10.1109/ICCV.2017.244
Zhu J, Zhang R, Pathak D, Darrell T, Efros A, Wang O, Shechtman E (2017b) Toward multimodal image-to-image translation. arXiv preprint. https://doi.org/10.48550/arXiv.1711.11586
Acknowledgements
The authors are grateful to Mengyang Lu (Academy for Engineering and Technology, Fudan University, China) for providing the experimental supports. This work was supported in part by the National Natural Science Foundation of China (61871263, 12274092, and 12034005), in part by the Explorer Program of Shanghai (21TS1400200), in part by the Natural Science Foundation of Shanghai (21ZR1405200), and in part by the Medical Engineering Fund of Fudan University (YG2022-6).
Author information
Authors and Affiliations
Contributions
XL and BL performed the analysis, drafted the paper. BL and CL supported in gathering study data, analysis design. XL and DT made critical revisions and approved the final version.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent to publication
Not applicable.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, X., Li, B., Liu, C. et al. Virtual Fluorescence Translation for Biological Tissue by Conditional Generative Adversarial Network. Phenomics 3, 408–420 (2023). https://doi.org/10.1007/s43657-023-00094-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43657-023-00094-1