Abstract
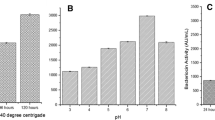
Response surface methodology (RSM) and artificial neural networks (ANN) are considered the most efficient way for optimization and modeling studies to design and develop various biosimilars. The primary objective of this study was to create empirical modeling and optimization of media parameters for producing B. halotolerans VSH 09 lipase using RSM and ANN. One-factor-at-a-time (OFAT) analysis revealed that triacylglycerols hydrolyzed by lipase manifest substantial activity. The subsequent screening for best carbon, nitrogen, and inducer was performed using the Placket–Burman design (PBD). The statistically significant variables were further examined for their optimum level using Box–Behnken design (BBD). The lipase production was optimized (26.04 IU/ml) under the ideal molasses (2.5%), peptone (2%), and salt (0.1% CaCO3, 0.1% (NH4)2SO4, and 0.1% MgSO4.7H2O). Both models revealed impeccable predictions; however, more interestingly, it was evaluated that ANN outperforms the RSM regarding data fitting and estimation capabilities.




Similar content being viewed by others
Availability of data and materials
Not applicable.
References
Basri M, Rahman RN, Ebrahimpour A, Salleh AB, Gunawan ER, Rahman MB. Comparison of estimation capabilities of response surface methodology (RSM) with artificial neural network (ANN) in lipase-catalyzed synthesis of palm-based wax ester. BMC Biotechnol. 2007;7:53.
Ebrahimpour A, Abd Rahman RN, Ean Ch’ng DH, Basri M, Salleh AB. A modeling study by response surface methodology and artificial neural network on culture parameters optimization for thermostable lipase production from a newly isolated thermophilic Geobacillus sp. strain ARM. BMC Biotechnol. 2008;8:96.
Lau H-L, Wong FWF, Rahman RNZRA, Mohamed MS, Ariff AB, Hii S-L. Optimization of fermentation medium components by response surface methodology (RSM) and artificial neural network hybrid with genetic algorithm (ANN–GA) for lipase production by Burkholderia cenocepacia ST8 using used automotive engine oil as substrate. Biocatal Agric Biotechnol. 2023;50:102696.
Khaleghi MK, Savizi ISP, Lewis NE. Shojaosadati SAJBJ: Synergisms of machine learning and constraint-based modeling of metabolism for analysis and optimization of fermentation parameters. Biotechnol J. 2021;16(11):2100212.
Sarmah N, Mehtab V, Bugata LSP, Tardio J, Bhargava S, Parthasarathy R, Chenna SJBT. machine learning aided experimental approach for evaluating the growth kinetics of Candida antarctica for lipase production. Bioresour Technol. 2022;352:127087.
Haider MA, Pakshirajan K, Singh A, Chaudhry S. Artificial neural network-genetic algorithm approach to optimize media constituents for enhancing lipase production by a soil microorganism. Appl Biochem Biotechnol. 2008;144(3):225–35.
Cai G, Zheng W, Yang X, Zhang B, Zheng T. Combination of uniform design with artificial neural network coupling genetic algorithm: an effective way to obtain high yield of biomass and algicidal compound of a novel HABs control actinomycete. Microb Cell Fact. 2014;13(1):75.
Bezerra MA, Santelli RE, Oliveira EP, Villar LS, Escaleira LA. Response surface methodology (RSM) as a tool for optimization in analytical chemistry. Talanta. 2008;76(5):965–77.
Salim N, Santhiagu A, Joji K. Process modeling and optimization of high yielding L-methioninase from a newly isolated Trichoderma harzianum using response surface methodology and artificial neural network coupled genetic algorithm. Biocatal Agric Biotechnol. 2019;17:299–308.
Gupta N, Sahai V, Gupta R. Alkaline lipase from a novel strain Burkholderia multivorans: statistical medium optimization and production in a bioreactor. Process Biochem. 2007;42(4):518–26.
Kumar P, Valsa A. Optimization of culture media and cultural conditions for the production of extracellular lipase by Bacillus coagulans. Indian J Biotechnol. 2007;6:114–7.
Hombalimath V, Udapudi B, Patil L, Shet A, Yaraguppi D, Tennalli G. Isolation and characterization of lipolytic microorganisms from oil contaminated soil. Int J Adv Eng Sci Technol (IJAEST). 2012;2:2012.
Nelofer R, Ramanan RN, Rahman RN, Basri M, Ariff AB. Comparison of the estimation capabilities of response surface methodology and artificial neural network for the optimization of recombinant lipase production by E. coli BL21. J Ind Microbiol Biotechnol. 2012;39(2):243–54.
Semache N, Fatiha B, Kerouaz B, Belhaj I, Bounour S, Belghith H, Ladjama A, Djeghaba Z. Artificial neural networks and response surface methodology approach for optimization of an eco-friendly and detergent-stable lipase production from actinomadura keratinilytica strain Cpt29. Acta Chim Slov. 2021;68:575–86.
Mahnashi M, Hombalimath V, Shaikh I, Muddapur U, Desai S, Achappa S, El-Sherbiny M, Ghoneim M, Jefri O, Alshahrani M, et al. Production of extracellular lipase by bacillus halotolerans from oil-contaminated soil in a pilot-scale submerged bioreactor. Processes. 2022;10:1548.
Sirisha E, Rajasekar N. Narasu MLJAiBR: Isolation and optimization of lipase producing bacteria from oil contaminated soils. Adv Biol Res. 2010;4(5):249–52.
Mandepudi D, Mandepudi B, Chowdary G, Gajbhiye A. Screening, isolation and biochemical characterization of novel lipase producing bacteria from soil samples. Int J Biol Eng. 2012;2:18–22.
Kumar A, Parihar SS, Batra N. Enrichment, isolation and optimization of lipase-producing Staphylococcus sp. from oil mill waste (oil cake). J Exp Sci. 2013;3(8):26–30.
Maia MM, Heasley A, Camargo de Morais MM, Melo EH, Morais MA Jr, Ledingham WM, Lima Filho JL. Effect of culture conditions on lipase production by Fusarium solani in batch fermentation. Bioresource Technol. 2001;76(1):23–7.
Wang J, Wan W. Optimization of fermentative hydrogen production process using genetic algorithm based on neural network and response surface methodology. Int J Hydrogen Energy. 2009;34(1):255–61.
Dalmau E, Montesinos JL, Lotti M, Casas C. Effect of different carbon sources on lipase production by Candida rugosa. Enzyme Microb Technol. 2000;26(9–10):657–63.
Aachapa S, Hombalimath VS, Kamaraddi JR, Desai SV, Shet AR, Patil LR. Patil JRJBBRC: Statistical optimization of lipase production from bacillus species by submerged fermentation. Biosci Biotechnol Res Com. 2021;14(1):264–9.
Kaushik R, Saran S, Isar J, Saxena RK. Statistical optimization of medium components and growth conditions by response surface methodology to enhance lipase production by Aspergillus carneus. J Mol Catal B Enzym. 2006;40:121–6.
Plackett RL, Burman JP. The design of optimum multifactorial experiments. Biometrika. 1946;33(4):305–25.
Salihu A, Bala M, Bala SM. Application of Plackett-Burman experimental design for lipase production by Aspergillus niger using shea butter cake. ISRN Biotechnol. 2013;2013: 718352.
Cavas L, Topcam G, Gundogdu-Hizliates C, Ergun Y. Neural network modeling of AChE inhibition by new carbazole-bearing oxazolones. Interdiscip Sci Comput Life Sci. 2019;11(1):95–107.
Ferreira SLC, Bruns RE, Ferreira HS, Matos GD, David JM, Brandão GC, da Silva EGP, Portugal LA, dos Reis PS, Souza AS, et al. Box-Behnken design: an alternative for the optimization of analytical methods. Anal Chim Acta. 2007;597(2):179–86.
Baş D, Boyacı İH. Modeling and optimization II: comparison of estimation capabilities of response surface methodology with artificial neural networks in a biochemical reaction. J Food Eng. 2007;78(3):846–54.
Dutta JR, Dutta PK, Banerjee R. Optimization of culture parameters for extracellular protease production from a newly isolated Pseudomonas sp. using response surface and artificial neural network models. Process Biochem. 2004;39(12):2193–8.
Singh P, Shera SS, Banik J, Banik RM. Optimization of cultural conditions using response surface methodology versus artificial neural network and modeling of l-glutaminase production by Bacillus cereus MTCC 1305. Biores Technol. 2013;137:261–9.
Chiranjeevi P, Pandian M, Thadikamala S. Integration of artificial neural network modeling and genetic algorithm approach for enrichment of laccase production in solid state fermentation by Pleurotus ostreatus. BioResources. 2014;9(2):2459–70.
Kotogán A, Furka ZT, Kovács T, Volford B, Papp DA, Varga M, Huynh T, Szekeres A, Papp T, Vágvölgyi C, et al. Hydrolysis of edible oils by fungal lipases: an effective tool to produce bioactive extracts with antioxidant and antimicrobial potential. Foods (Basel, Switzerland). 2022;11(12):1711.
Ghosh PK, Saxena RK, Gupta R, Yadav RP, Davidson S. Microbial lipases: production and applications. Sci Progress. 1996;79(2):119–57.
Mahler GF, Kok RG, Cordenons A, Hellingwerf KJ, Nudel BC. Effects of carbon sources on extracellular lipase production and lipA transcription in Acinetobacter calcoaceticus. J Ind Microbiol Biotechnol. 2000;24(1):25–30.
Tembhurkar V, Kulkarni M, Peshwe SA. Optimization of lipase production by Pseudomonas spp in submerged batch process in shake flask culture. Sci Res Reporter. 2012;2(1):46–50.
Gupta R, Gupta N, Rathi P. Bacterial lipases: an overview of production, purification and biochemical properties. Appl Microbiol Biotechnol. 2004;64(6):763–81.
Alhelli AM, Abdul Manap MY, Mohammed AS, Mirhosseini H, Suliman E, Shad Z, Mohammed NK, Meor Hussin AS. Use of response surface methodology for partitioning, one-step purification of alkaline extracellular lipase from Penicillium candidum (PCA 1/TT031). J Chromatogr B. 2016;1039:66–73.
Javed M, Khan T, Haq I. A 23 level full factorial design to optimize cultural conditions for lipase production by consortium of aspergillus niger and trichoderma viride. Res J Microbiol. 2007;2:639–44.
Gunawan ER, Basri M, Rahman MBA, Salleh AB, Rahman RNZA. Study on response surface methodology (RSM) of lipase-catalyzed synthesis of palm-based wax ester. Enzyme Microb Technol. 2005;37(7):739–44.
Senanayake J, Shahidi F. Lipase-catalyzed incorporation of docosahexaenoic acid (DHA) into borage oil: optimization using response surface methodology. Food Chem. 2002;77:115–23.
Giordano PC, Martínez HD, Iglesias AA, Beccaria AJ, Goicoechea HC. Application of response surface methodology and artificial neural networks for optimization of recombinant Oryza sativa non-symbiotic hemoglobin 1 production by Escherichia coli in medium containing byproduct glycerol. Biores Technol. 2010;101(19):7537–44.
He Y-Q, Tan T-W. Use of response surface methodology to optimize culture medium for production of lipase with Candida sp. 99-125. J Mol Catal B Enzym. 2006;43(1):9–14.
Lima C, Fontes Coelho L, Contiero J. The use of response surface methodology in optimization of lactic acid production: focus on medium supplementation, temperature and pH control. Food Technol Biotechnol. 2010;48(2):175–81.
Ruchi G, Anshu G, Khare SK. Lipase from solvent tolerant Pseudomonas aeruginosa strain: production optimization by response surface methodology and application. Biores Technol. 2008;99(11):4796–802.
Manohar B, Divakar S. An artificial neural network analysis of porcine pancreas lipase catalysed esterification of anthranilic acid with methanol. Process Biochem. 2005;40(10):3372–6.
Whiteman JK, Gueguim Kana EB. Comparative assessment of the artificial neural network and response surface modelling efficiencies for biohydrogen production on sugar cane molasses. BioEnergy Research. 2014;7(1):295–305.
Wang X, Jin B. Process optimization of biological hydrogen production from molasses by a newly isolated Clostridium butyricum W5. J Biosci Bioeng. 2009;107(2):138–44.
Prakasham RS, Sathish T, Brahmaiah P. Imperative role of neural networks coupled genetic algorithm on optimization of biohydrogen yield. Int J Hydrogen Energy. 2011;36(7):4332–9.
Sewsynker-Sukai Y, Kana G, Lateef A. Modelling of biohydrogen generation in microbial electrolysis cells (MECs) using a committee of artificial neural networks (ANNs). Biotechnol Biotechnol Equip. 2015;29(6):1208–15.
Acknowledgements
The authors would like to thank all the support received from KLE Technological University and Amity University Uttar Pradesh. A special thanks to Dr. Gururaj Pathak, Assistant Professor, Kirloskar Institute of Management, Harihara, for his constant support and coordination during manuscript preparation.
Funding
No source of funding has been received for the current research work.
Author information
Authors and Affiliations
Contributions
VH and DMG contributed equally to the experimental design and execution of results. VH was involved in data compiling, and DMG extensively contributed to manuscript writing and formatting.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Consent is applicable from both authors.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hombalimath, V.S., Gurumurthy, D.M. Response surface methodology (RSM) and artificial neural network (ANN) integrated optimization for lipase production by Bacillus holotolerans. Syst Microbiol and Biomanuf (2023). https://doi.org/10.1007/s43393-023-00220-0
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s43393-023-00220-0