Abstract
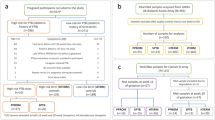
Biomarkers associated with spontaneous preterm birth (sPTB) before labor onset could aid in prediction, triage, and stratification for testing interventions. In this study we examined maternal blood EBF1-correlated long non-coding RNAs (lncRNAs) in relation to sPTB. We retrieved all lncRNA transcripts from a public gene expression dataset (GSE59491) derived from maternal blood in trimesters 2 and 3 from a Canadian cohort with a matched set of sPTB (n = 51) and term births (n = 106). LncRNA transcripts differentially expressed (limma moderated t-tests) in sPTB vs. term were tested for correlations (Pearson) with EBF1 mRNA levels in the same blood samples. Using logistic regression, EBF1-correlated lncRNAs were divided into tertiles and assessed in relation to odds of sPTB. Two lncRNA transcripts in the 3rd trimester maternal blood were differentially expressed between sPTB and term births (all p < 0.001 and FDR < 0.250) and positively and negatively correlated with EBF1 mRNA levels. They were as follows: (1) LINC00094 r = 0.196 (95% CI: 0.039 to 0.344), p = 0.015, and BH adjusted p = 0.022 and (2) LINC00870 r = − 0.303 (95% CI: − 0.441 to − 0.152), p < 0.001, and BH adjusted p < 0.001. As compared with term births, sPTBs were more likely to be in the highest tertile of LINC00870 (odds ratio (OR) = 4.08 (95% CI 1.60, 10.40), p = 0.003) and the lowest tertile of LINC00094 (OR = 5.16 (95% CI 1.96, 13.61), p < 0.001). Two sPTB-associated EBF1-correlated lncRNAs (LINC00870 and LINC00094) had multiple potential enhancers containing EBF1 binding site(s). Our current findings, along with previous reports linking EBF1 and sPTB, motivate additional research on the EBF1 gene-related gene expression and regulation in relation to sPTB within other cohorts and within laboratory-based models.

Similar content being viewed by others
References
Goldenberg RL, Culhane JF, Iams JD, Romero R. Epidemiology and causes of preterm birth. Lancet. 2008;371:75–84.
Manuck TA, Esplin MS, Biggio J, Bukowski R, Parry S, Zhang H, et al. The phenotype of spontaneous preterm birth: application of a clinical phenotyping tool. Am J Obstet Gynecol. 2015;212(4):487.e1–487.e11.
Heng YJ, Pennell CE, McDonald SW, Vinturache AE, Xu J, Lee MWF, et al. Maternal whole blood gene expression at 18 and 28 weeks of gestation associated with spontaneous preterm birth in asymptomatic women. PLoS One. 2016;11(6):e0155191.
Esplin MS, Elovitz MA, Iams JD, Parker CB, Wapner RJ, Grobman WA, et al. Predictive accuracy of serial transvaginal cervical lengths and quantitative vaginal fetal fibronectin levels for spontaneous preterm birth among nulliparous women. JAMA. 2017;317(10):1047–56.
Norman JE, Shennan AH. Prevention of preterm birth e why can't we do any better? Lancet. 2012;381(9862):184e5.
Zhang G, Feenstra B, Bacelis J, Liu X, Muglia LM, Juodakis J, et al. Genetic associations with gestational duration and spontaneous preterm birth. N Engl J Med. 2017;377(12):1156–67.
Nechanitzky R, Akbas D, Scherer S, Gyory I, Hoyler T, Ramamoorthy S, et al. Transcription factor EBF1 is essential for the maintenance of B cell identity and prevention of alternative fates in committed cells. Nat Immunol. 2013;14:867–75.
Zhou G, Holzman C, Heng YJ, Kibschull M, Lye SJ, Vazquez A. EBF1 gene mRNA levels in maternal blood and spontaneous preterm birth. Reprod Sci. 2020;27:316–24.
Zhou G, Holzman C, Heng YJ, Kibschull M, Lye SJ. Maternal blood EBF1-based microRNA transcripts as biomarkers for detecting risk of spontaneous preterm birth: a nested case-control study. J Matern Fetal Neonatal Med. 2020:1–9. https://doi.org/10.1080/14767058.2020.1745178.
Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–89.
Ransohoff JD, Wei Y, Khavari PA. The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol. 2018;19(3):143–57.
He X, He Y, Xi B, Zheng J, Zeng X, Cai Q, et al. LncRNAs expression in preeclampsia placenta reveals the potential role of LncRNAs contributing to preeclampsia pathogenesis. PLoS One. 2013;8(11):e81437.
Luo X, Li X. Long non-coding RNAs serve as diagnostic biomarkers of preeclampsia and modulate migration and invasiveness of trophoblast cells. Med Sci Monit. 2018;24:84–91.
Shi Z, Zhao C, Long W, Ding H, Shen R. Microarray expression profile analysis of long non-coding RNAs in umbilical cord plasma reveals their potential role in gestational diabetes-induced macrosomia. Cell Physiol Biochem. 2015;36(2):542–54.
Romero R, Tarca AL, Chaemsaithong P, Miranda J, Chaiworapongsa T, Jia H, et al. Transcriptome interrogation of human myometrium identifies differentially expressed sense-antisense pairs of protein-coding and long non-coding RNA genes in spontaneous labor at term. J Matern Fetal Neonatal Med. 2014;27(14):1397–408.
Luo X, Shi Q, Gu Y, Pan J, Hua M, Liu M, et al. LncRNA pathway involved in premature preterm rupture of membrane (PPROM): an epigenomic approach to study the pathogenesis of reproductive disorders. PLoS One. 2013;8(11):e79897.
Luo X, Pan J, Wang L, Wang P, Meijiao Zhang M, Liu M, et al. Epigenetic regulation of lncRNA connects ubiquitin-proteasome system with infection-inflammation in preterm births and preterm premature rupture of membranes. BMC Pregnancy Childbirth. 2015;15:35. https://doi.org/10.1186/s12884-015-0460-0.
Zhao X, Dong X, Luo X, Pan J, Ju W, Zhang M, et al. Ubiquitin-proteasome-collagen (CUP) pathway in preterm premature rupture of fetal membranes. Front Pharmacol. 2017;8:310. https://doi.org/10.3389/fphar.2017.00310.
Law CW, Alhamdoosh M, Su S, Smyth GK, Ritchie ME. RNA-seq analysis is easy as 1–2-3 with limma, Glimma and edgeR. F1000Research. 2016;5:1408.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. https://doi.org/10.1093/nar/gkv007.
Wang Y, Jiang T, Li Z, Lu L, Zhang R, Zhang D, et al. Analysis of differentially co-expressed genes based on microarray data of hepatocellular carcinoma. Neoplasma. 2017;64(2):216–21.
Drizik E, Corbett S, Zheng Y, Vermeulen R, Dai Y, Hu W, et al. Transcriptomic changes in the nasal epithelium associated with diesel engine exhaust exposure. Environ Int. 2020;137:105506. https://doi.org/10.1016/j.envint.2020.105506.
Bishara AJ, Hittner JB. Confidence intervals for correlations when data are not normal. Behav Res Methods. 2017;49:294–309.
Hu X, Jung A, Qin G. Interval estimation for the correlation coefficient. Am Stat. 2018;74:29–36. https://doi.org/10.1080/00031305.2018.1437077.
Curtin F, Schulz P. Multiple correlations and Bonferroni's correction. Biol Psychiatry. 1998;44(8):775–7.
Yu H, Liu BH, Ye ZQ, Li C, Li YX, Li YY. Link-based quantitative methods to identify differentially coexpressed genes and gene pairs. BMC Bioinformatics. 2011;12:315.
Revelle W. psych: Procedures for psychological, psychometric, and personality research. Northwestern University, Evanston, Illinois. R package version 1.9.12. 2019; https://CRAN.R-project.org/package=psych. Accessed 15 Oct 2019.
Devika S, Jeyaseelan L, Sebastian G. Analysis of sparse data in logistic regression in medical research: a newer approach. J Postgrad Med. 2016;62(1):26–31.
Davison AC, Hinkley DV. Bootstrap methods and their applications. Cambridge: Cambridge University Press; 1997. ISBN 0-521-57391-2
Canty A, Ripley B. boot: Bootstrap R (S-Plus) Functions. R package version 1.3–23. 2019.
Fishilevich S, Nudel R, Rappaport N, Hadar R, Plaschkes I, Iny Stein T, et al. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database (Oxford). 2017;2017:bax028. https://doi.org/10.1093/database/bax028.
Stelzer G, Rosen R, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, et al. The GeneCards Suite: from gene data mining to disease genome sequence analysis. Curr Protoc Bioinformatics. 2016;54:1.30.1–1.30.33. https://doi.org/10.1002/cpbi.5.
Kamburov A, Stelzl U, Lehrach H, Herwig R. The ConsensusPathDB interaction database: 2013 update. Nucleic Acids Res. 2013;41:D793–800.
Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5:e13984.
Ullah U, Tripathi P, Lahesmaa R, Rao KV. Gene set enrichment analysis identifies LIF as a negative regulator of human Th2 cell differentiation. Sci Rep. 2012;2:464.
Lukin K, Fields S, Guerrettaz L, Straign D, Rodriguez V, Zandi S, et al. A dose-dependent role for EBF1 in repressing non-B-cell-specific genes. Eur J Immunol. 2011;41:1787–93.
Jeney V, Balla J, Yachie A, Varga Z, Vercellotti GM, Eaton JW, et al. Pro-oxidant and cytotoxic effects of circulating heme. Blood. 2002;100:879–87.
Chiabrando D, Vinchi F, Fiorito V, Mercurio S, Tolosano E. Heme in pathophysiology: a matter of scavenging, metabolism and trafficking across cell membranes. Front Pharmacol. 2014;5:1–24.
Vasconcellos LR, Dutra FF, Siqueira MS, Paula-Neto HA, Dahan J, Kiarely E, et al. Protein aggregation as a cellular response to oxidative stress induced by heme and iron. Proc Natl Acad Sci U S A. 2016;113(47):E7474–82.
Ferrer MD, Mestre-Alfaro A, Martínez-Tomé M, Carrera-Quintanar L, Capó X, Jiménez-Monreal AM, et al. Haem biosynthesis and antioxidant enzymes in circulating cells of acute intermittent porphyria patients. PLoS One. 2016;11(10):e0164857.
Maitra D, Bragazzi Cunha J, Elenbaas JS, Bonkovsky HL, Shavit JA, Omary MB. Porphyrin-induced protein oxidation and aggregation as a mechanism of porphyria-associated cell injury. Cell Mol Gastroenterol Hepatol. 2019;8(4):535–48.
Moore TA, Ahmad IM, Zimmerman MC. Oxidative stress and preterm birth: an integrative review. Biol Res Nurs. 2018;20(5):497–512.
Lynch AM, Wagner BD, Deterding RR, Giclas PC, Gibbs RS, Janoff EN, et al. The relationship of circulating proteins in early pregnancy with preterm birth. Am J Obstet Gynecol. 2016;214(4):517.e1–8.
D'Silva AM, Hyett JA, Coorssen JR. Proteomic analysis of first trimester maternal serum to identify candidate biomarkers potentially predictive of spontaneous preterm birth. J Proteome. 2018;178:31–42.
Author information
Authors and Affiliations
Contributions
G.Z. developed research ideas, conducted data analyses including statistical analysis and bioinformatics analysis, interpreted the data, and drafted manuscript. C.H. provided advice on developing research ideas and conducting data analyses, interpreted the data, as well as reviewed and revised manuscript. B.C., P.W., Y.J.H, M.K., S.J.L., and E.K. reviewed and revised manuscript. All authors gave final approval of the version to be published.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Ethics Approval
Not applicable.
Informed Consent
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhou, G., Holzman, C., Chen, B. et al. EBF1-Correlated Long Non-coding RNA Transcript Levels in 3rd Trimester Maternal Blood and Risk of Spontaneous Preterm Birth. Reprod. Sci. 28, 541–549 (2021). https://doi.org/10.1007/s43032-020-00320-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43032-020-00320-5