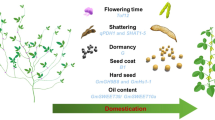
Abstract
Genetic diversity is a cornerstone of crop improvement, However, cultivated soybean (Glycine max) has undergone several genetic bottlenecks, including domestication in China, the introduction of landraces to other areas of the world and, latterly, selective breeding, leading to low genetic diversity the poses a major obstacle to soybean improvement. By contrast, there remains a relatively high level of genetic diversity in soybean’s wild relatives, especially the perennial soybeans (Glycine subgenus Glycine), which could serve as potential gene pools for improving soybean cultivars. Wild soybeans are phylogenetically diversified and adapted to various habitats, harboring resistance to various biotic and abiotic stresses. Advances in genome and transcriptome sequencing enable alleles associated with desirable traits that were lost during domestication of soybean to be discovered in wild soybean. The collection and conservation of soybean wild relatives and the dissection of their genomic features will accelerate soybean breeding and facilitate sustainable agriculture and food production.
Similar content being viewed by others
Introduction
Climate change, coupled with an increasing human population, is expected to lead to global food shortages, especially for protein-rich foods. To feed a growing population of over 9 billion people by 2050, the current rate of agricultural productivity will need to increase twofold (Tilman et al. 2011; Ray et al. 2013). Thus, developing crops with higher yields has become an urgent task for scientists. The cultivated soybean [Glycine max (L.) Merr.], which is rich in both protein and oil, is one of the most important crop in the word (Li et al. 2008), providing ~ 50% of the word’s oilseed production (www.fao.org).
Soybean originated in China and is thought to have been domesticated from its wild progenitor Glycine soja Sieb. & Zucc. ~ 5000 years ago (Hymowitz, 1970; Li et al., 2008). Although abundant soybean germplasm has been collected, much of its genetic diversity has been lost during domestication, during which resulted in the production of numerous Asian landraces of soybean. More recently, selective breeding has been used to meet human needs, giving rise to ‘genetic bottlenecks’ (Hyten et al. 2006). A study by Gai and Zhao (2001) determined that 38 ancestral varieties represent 54.18% of the nuclear genetic material of the 651 soybean cultivars released in China between 1923 and 1995. The situation is even worse in North America, one of the world’s largest soybean producing areas, where soybean genetic diversity of soybean is extremely low because of founding events. Of the 45,000 unique Asian landraces collected worldwide, 80 account for 99% of the collective parentage of North American soybean cultivars released between 1947 and 1988 (Carter et al. 2004) and 79% of rare alleles (frequency < 0.10) present in the Asian landraces have been lost (Hyten et al. 2006) in landraces introduced to North America. Compared with its annual wild relative G. soja Sieb. & Zucc., cultivated soybean has lost ~ 50% of its sequence diversity (Hyten et al. 2006). Such low genetic diversity of domesticated germplasm not only hinders current soybean breeding and improvement efforts, but also makes this important crop vulnerable to emerging biotic and abiotic stressors, thus threatening long-term food security (Tanksley and McCouch 1997).
The use of crop wild relatives of crop in traits improvements, particularly for increased pest and disease resistance, has been successful in tomato, barley, and wheat (Zhang and Batley 2020). Likewise, to overcome the bottleneck effect of cultivation in soybean, researchers have sought to broaden its gene pool using wild relatives. Harland and de Wet (1971) defined three main types of gene pools (GP), based on the success rate of hybridization among species (Fig. 1). GP-1 contains germplasm that can be easily hybridized, and includes all soybean cultivars, landraces, and its wild progenitor G. soja Sieb. & Zucc. GP-2 comprises species that can be crossed with GP-1 without causing F1 infertility (Dwivedi et al. 2008); however, to date, there are no soybean species in this group. GP-3 comprises 26 wild perennial species, considered to be confined to Australia. These serve as the outer limits for potential gene pools, since F1 hybridization seeds produced by crossing GP-3 species and soybean require in vitro techniques to rescue lethality (Singh et al. 2017).
Classification of gene pools for the cultivated soybean based on Harlan and de Wet (1971)
Compared with cultivated soybean, wild soybean relatives have weedy, prostrate growth habits and exhibit distinct agronomic traits, such as smaller seed size, and increased pod number and seed number per plant (Hymowitz 1970; Singh et al. 2007; Singh and Hymowitz 2011). They also usually have a higher reproductive rate and increased tolerance to both biotic and abiotic stresses. Identifying the genes and alleles that are specific to wild germplasm and associated with agronomic traits is key for applying wild germplasm to broaden the genetic background and accelerate molecular breeding in soybean. Obtaining intact and accurate genomic information for wild germplasm is essential for tracking genomic variations, identifying quantitative trait loci (QTL), and conducting association studies (Xie et al. 2019).
A reference genome for the cultivated soybean accession ‘Williams 82’ was published in 2010 (Schmutz et al. 2010) and reference genomes for its annual relative G. soja accessions IT182932 and W05 was published by Kim et al (2011) and Xie et al. (2019), respectively. Thanks to the recent, rapid development of genome sequencing technology, and the reduction in sequencing costs, more pan-genome analyses have been conducted for both cultivated soybean plants and their wild relatives, providing abundant genome resources for soybean improvement. Here, we summarize advances in using wild soybeans as valuable resources in soybean breeding efforts, and highlight recent genomic accomplishments as well as valuable genes (Table 1) identified in both annual and perennial wild soybean species.
Genetic architecture of soybean’s wild relatives
The genomes of Glycine-grouped species were classified into seven groups based on their ability to produce fertile hybrids, and the degree to which meiotic chromosomes pair (Sherman-Broyles et al. 2014). The annual wild relative of soybean (Glycine soja Sieb. & Zucc.) and cultivated soybean (G. max (L.) Merr.) both belong to the G genome group and have the same number of chromosomes (2n = 40). The first sequenced genome of G. soja was reported by Kim et al. (2010) for G. soja ‘IT182932’. Illumina short reads were used to produce a genome representing 97.65% coverage of the published G. max ‘Williams 82’ genome sequence. Comparative analysis of the two genomes indicated ~ 0.31% sequence variation, including single nucleotide substitutions and small insertion/deletions.
To better characterize the genomic content and evolutionary history of G. soja, Lam et al., (2010) sequenced 31 wild and cultivated soybean accessions at ~ 5 × coverage and identified 205,614 single nucleotide polymorphisms (SNPs) for QTL mapping and association studies, as well as marker-assisted soybean breeding. Later, Li et al. (2014) sequenced and de novo-assembled seven representative accessions with an average of > 100 × Illumina reads, and constructed a pan-genome for G. soja. Compared with the single accession sequenced, the pan-genome was 30.2 Mbp larger and covered 94% of the annotated assembly for G. max ‘Williams 82’ (v1). Besides identifying core genes shared by all seven accessions, Li and colleagues reported a set of dispensable genes exhibiting a higher rate of variation, presumably as a result of adaptation to diverse environments (Li et al. 2014).
The first reference-grade genome of G. soja was constructed by Xie et al. (2019) for the accession G. soja ‘W05’. This used a combination of Illumina short reads and PacBio long reads, and enabled the identification of large structural variations and gene copy number variations. Through genome-wide comparison of the wild species ‘W05’ and the cultivated soybean accession ‘Williams 82’, they successfully identified an inversion associated with seed coat color during domestication, thus demonstrating the importance of high-quality genome assembly. In 2020, a milestone was reached on the way to unraveling the species-wide genetic diversity of annual soybeans, with the publication of the complete genome sequences for 26 representative accessions. In addition, 2898 accessions were deeply re-sequenced, including 103 annual wild soybeans (Liu et al. 2020). Comparison of annual wild relatives to cultivated soybeans identified several structural variations and candidate genes associated with domestication, such as seed coat pigmentation, thus providing valuable resources for future soybean breeding.
The subgenus Glycine contains ~ 30 perennial species indigenous to Australia (Sherman-Broyles et al. 2014) that are geographically and reproductively isolated from G. max and G. soja. Compared with the annual wild relatives of soybean, the genomes of its perennial wild relatives are much more complex, belonging to six different genome groups (A–F). Unlike the subgenus Soja, members of the perennial subgenus Glycine have 2n = 38, 40, 78, and 80 chromosomes. Except for G. tomentella (2n = 78), hybrids of the subgenera Glycine and Soja are infertile, which making their application in conventional soybean breeding challenging. Therefore, much effort has been directed toward analyzing the annual relatives of soybean, even though many valuable agronomically related traits have been identified in the perennial species. However, this limitation could be resolved via the rapid progress of biotechnology, including improved genetic transformation and cutting-edge genome editing methods (e.g., Kim et al. 2017; Fig. 2). After the release of the genome sequences of various G. soja accessions, genetic analysis revealed lower heterozygosity in G. soja than expected, even for selfing species (Sherman-Broyles et al. 2014; Guo et al. 2012; He et al. 2012). Hyten et al. (2007) also reported that the genetic diversity of G. soja is approximately twofold to fivefold lower than that of the wild relatives of other plants, such as the wild brassica Arabidopsis thaliana (Schmid et al., 2005), wild barley (Morrell et al., 2005), and wild maize relatives (teosinte) (Wright et al., 2005), the genetic diversity of G. soja is about twofold to fivefold lower. Thus, the genetic diversity of cultivated soybean is not only depauperate because of domestication; but the bottleneck effect also seems to have occurred in its annual wild relatives following their speciation. In contrast, perennial wild soybeans are more phylogenetically diversified and adapted to various habitats. Therefore, identifying the genes underlying traits of interest in those species will be useful for the genetic modification of cultivated soybean.
Although efforts have been made to explore the genomic features of perennial soybeans, their genomic analysis is far behind that of their annual relatives and has been focused mainly on relatively small genomic segments because of the lack of complete reference sequences. The small segments studied to date include a 1-Mbp syntenic region surrounding the Rpg-1b locus on chromosome 13 (Innes et al. 2008; Wawrzynski et al. 2008), and 6–350 kb regions studied in the SoyMapII project. The first whole genome assembly of the subgenus Glycine was reported by Liu et al. (2018). They sequenced and assembled the G. latifolia genome using 10× Genomics linked-reads, representing 83.2% of the estimated genome. Synteny analysis found high genome collinearity for 12 chromosomes, while large rearrangements were identified for the other eight chromosomes. Analysis of gene families identified 3,148 G. latifolia-specific gene families, and genes related to defense responses, such as NB-ARC-encoding genes, were found to be overrepresented, presumably contributing to its excellent disease resistance. We recently sequenced and constructed an almost-completed reference genome for five perennial Glycine species, including one representative species for each genome group A–D and F, and built a pan-genome for the subgenus Glycine. Combining our data with the 26 pan-genomes of annual soybean constructed by Liu et al. (2020), we identified ~ 5000 core genes shared by all five perennial Glycine species. We also identified more than 60,000 non-core genes specific to the subgenus Glycine, which were either deleted or pseudolized in the subgenus Soja after they split from their common ancestors. An analysis of the ratio of nonsynonymous to synonymous changes (Ka/Ks) revealed a considerable number of genes showing signs of adaptive evolution. These genes are likely to be associated with the unique features of the subgenus Glycine, thus serving as potential genetic resources for soybean improvement.
Beneficial traits of wild soybean relatives
Biotic stress resistance
Soybean cyst nematode (SCN, caused by the pathogen Heterodera glycines) is the most devastating pathogen threatening soybean production, causing ~ 2.2% yield losses in Argentina, Brazil, and the America (Hartman 2015; Niblack and Riggs 2015; Hartman et al. 2017). Two major QTLs for breeding SCN-resistant soybean have been identified in G. max: rhg1 on chromosome 18 (Melito et al. 2010), and Rhg4, located on chromosome 8 (Concibido et al. 2004) are two major QTLs for breeding SCN-resistant soybean. However, nematodes have rapidly adapted to overcome the resistance provided by these two loci. To identify novel genes conferring SCN resistance, soybean breeders have screened a large portion of G. soja accessions for novel SCN resistance loci. Through QTL mapping and association analysis has identified SNPs associated with SCN infection, as well as two additional QTLs, located on chromosome 15 (cqSCN-006) and chromosome 18 (cqSCN-007) (Kim et al. 2011; Zhang et al. 2016, 2017c; Kofskyet al. 2018). Cumulatively, these alleles could provide a longer duration of SCN resistance than individual alleles.
Since the perennial soybean species originated from diverse geographical areas in Australia, they may possess a broader spectrum of SNC resistance. Several studies have examined SCN resistance in perennial soybean. Riggs and Schmitt (1998) evaluated nine perennial species; all had excellent SNC resistance to three different isolates of Heterodera glycines (HG), with little cyst production. Bauer et al. (2007) investigated 12 perennial soybeans for HG Type 0 resistance and identified immune responses in all species except G. curvata and G. pindacia. Wen et al. (2017) further evaluated 223 plant introductions (PIs) of G. tomentella and 59 PIs of 12 other perennial Glycines for resistance to HG Type 0, HG Type 2, and HG Type 1.2.3. The authors also evaluated 36 PIs for their resistance to HG Type 1.2.3.4.5.6.7, a population that can overcome all known soybean resistance genes. Of the 223 G. tomentella PIs, 177 showed resistance to one HG type. Of the 36 PIs challenged with HG Type 1.2.3.4.5.6.7, 35 were resistant and 16 PIs were totally immune. Herman et al. (2020) evaluated 16 accessions of G. argyrea, G. clandestine, G. dolichocarpoa, and G. tomentella for resistance to HG. All of these accessions were classified as resistant, and none of the G. tomentella accessions produced any cysts. Thus, the perennial Glycine spp. represent valuable resources for the discovery of novel gene loci that provide broad-spectrum SCN resistance to enhance soybean breeding.
Besides SCN resistance, wild soybean species are also rich resources of genes conferring resistance to sap-sucking insects (Zhang et al. 2019). Two QTL, Rag3b/c and Rag6 (Zhang et al., 2013; Zhang et al., 2017a, b), have been identified in G. soja (Lee et al., 2015) as contributing to soybean aphid (Aphis glycines) resistance, and one gene, Raso2 contributes to resistance to foxglove aphid (Aulacorthum solani) resistance. Screens of perennial Glycine spp. also revealed remarkable resistance to the leaf rust fungus Phakopsora pachyrhizi (Burdon and Marshall 1981; Hartman et al. 1992; Herman et al. 2020), brown spot (Lim and Hymowitz 1987), Phytophthora root rot (Kenworthy 1989), powdery mildew (Mignucci and Chamberlain 1978), soybean rust (Hartman et al. 1992; Hymowitz 1995), Sclerotinia stem rot, and sudden death syndrome (Hartman et al. 2000). Future research should explore the molecular mechanisms underlying multiple disease resistance traits by phenotyping wild soybean species, developing associated molecular markers, and identifying the genes that contribute to the observed resistance.
Abiotic stress tolerance
Drought reduces the productivity of soybean by up to 40% (Le et al. 2012). G. soja is commonly found in habitats with high water availability, and is thus usually not considered to be ideal material for developing drought-tolerant soybean varieties. Nonetheless, yield loss under drought stress was lower in transgenic G. max lines heterologously expressing GsWRKY20 from G. soja than in the corresponding non-transgenic control plants (Ning et al. 2017). By contrast, members of the subgenus Glycine are well adapted to drought conditions (Song et al. 2016).
Kao and Tsai (1998) evaluated the water use efficiency (WUE) of one annual wild relative, G. soja, and two perennial wild relative species, G. tomentella and G. tabacina. Of these three species tested, G. soja showed the lowest WUE, while G. tabacia exhibited the highest, consistent with their geographic distributions (the selected G. soja accessions lived in wet habitats, while G. tabacina was collected from an arid area). Morphologically, G. tabacina also showed the least vertical midday leaflet angles and lowest photosynthetic sensitivity to water deficiency. Reference genomes are now available for all three of these species. Transcriptome profiling and comparison among these species would help us to identify drought stress responsive genes with potential use in breeding drought-tolerant soybean.
Soil salinization is a global issue directly affects arable lands and causes crop losses. Salt-affected soils currently account for 8% of the world’s total land area (www.fao.org). Soybean is classified as a moderately salt-sensitive crop (Munns and Tester 2008). Although germplasms with varying levels of salt tolerance have been reported (Phang et al. 2008), and the yields of sensitive soybean cultivars reduced dramatically under salt stress (Lee et al. 2009; Katerji et al. 2003). Although reverse genetics approaches have revealed many soybean genes with functions in the salt stress response, only one major QTL, located on chromosome 3, has been repeatedly identified in different populations. A gene named GmSALT3 underlies this cloned QTL (Guan et al. 2014). Transcriptome analysis of different soybean varieties under salt stress suggested that the salt tolerance mechanism is genotype-specific. Peng et al. (2013) compared the physiological mechanisms of the G. soja accession ‘BB52’ with cultivated soybean ‘ZH13’, and established that the leaves of ‘BB52’ had higher relative water contents and water potentials than those of ‘ZH13’. Using annual wild relatives, several genes have been identified as contributing to salt tolerance, including Ncl2 (Lee et al. 2009), GmCHX1 (Qi et al. 2014), GmTIP2 (Zhang et al. 2017c), and GsGST (Ji et al. 2010). Both diploid and polyploid perennial wild relatives, particularly G. argyrea, G. clandestine, G. microphylla, G. tabacina, G. dolichocarp and G. tomentella, have higher salt tolerance than even the most salt-tolerant cultivated soybean (Pantalone et al. 1997; Kao et al. 2006; Lenis et al. 2011). The molecular mechanism underlying the outstanding salt tolerance of perennial wild relative species remains unclear. The availability of the G. tomentella reference genome will facilitate the identification of genes contributing to salt tolerance in perennial wild relatives.
Yield-related traits
Yield was the most important agronomic trait under selection during soybean domestication. In this respect, G. max is superior to its wild relatives. However, wild relatives of crops have been used for yield improvement in multiple species. For example, leaf angle is one of the most important agronomic traits contributing to improved growth densities and yield. Tian et al. (2019) created recombinant inbred lines between maize (Zea mays) and its wild ancestor species teosinte (Zea mexicana) and identified a gene named UPA2 that regulates leaf angle in maize. The authors identified the teosinte version of UPA2 and determined that it made leaves more upright when introduced into cultivated maize (Tian et al. 2019). In soybean, Akpertey et al. (2018) have reported that genetic introgression from perennial G. tomentella to cultivated G. max significantly increased seed yield. Other traits related to yield in genetically diverse accessions could also potentially be used in soybean breeding programs.
Photosynthetic efficiency is associated with crop yield, particularly when other factors such as salinity and cadmium stress are involved. Kao et al. (2003) evaluated photosynthetic gas exchange and chlorophyll a fluorescence in three wild soybeans (G. soja) and two perennial Glycine species (G. tomentella and G. tabacina) under salt stress. Different levels of sensitivity were observed among the three lines: photosynthesis was less affected by salt stress in the perennial Glycine spp. than in G. soja and was least affected in G. tomentella. Chen et al. (2013) and Xue et al. (2014) reported that the halophytic wild soybean ‘Dongying’ (G. soja Sieb. & Zucc. ‘ZYD 03262’) maintained higher photosynthetic activity under salt stress than cultivated soybean. Furthermore, Xue et al. (2014) determined that under salt stress, G. soja accumulated more Na+ in roots, but much less in leaves, than various G. max accessions. Thus, photosynthetic activity, stomatal conductance, and carboxylation efficiency were less affected by salt stress in G. soja than in the G. max accessions. The ability to maintain a steady leaf cell state might play a key role in the different salt stress responses of these accessions. Enhanced capacities for photosynthetic electron transport and for reversible forms of photoprotection have been reported in perennial allopolyploid Glycine spp. (Coate et al. 2012, 2013; Ilut et al. 2012); these advantages are presumably associated with their high ploidy levels. However, the molecular mechanisms underlying the enhanced photosynthetic ability in wild soybean remain to be elucidated.
The regulation of photoperiodic flowering is another important agronomic trait. Photoperiodic flowering is critical for regional adaptation and yield (Lin et al 2021), since early maturity is usually accompanied by low yields. Cultivated soybean and its annual wild relative G. soja are typical short-day plants that flower when the daylength is shorter than a certain threshold (Sedivy et al. 2017). Soybean cultivars are adapted for production in narrow bands of latitude (Kenworthy et al. 1989; Ohigashi et al. 2019). Varieties adapted to high latitude will bloom early when grown at low latitudes; such plants are normally short, with few pods. By contrast, varieties adapted to low latitude areas normally show delayed flowering, which prolongs vegetative growth for maximum yield potential; when grown at high latitudes, such plants cannot complete their lifecycles before winter (Lin et al. 2021). Combinations of forward and reverse genetic approaches have identified at least 18 major loci that participate in the photoperiod response in soybean, designated E1 to E11, J, Tof5, Tof11, Tof12, Tof18, Tof1111 and Tof112 (Xia et al. 2012; Watanabe et al. 2009, 2011; Liu et al. 2008; Cober 2011; Cober and Voldeng 2001; Kong et al. 2010; Zhai et al. 2014; Wang et al. 2019; Lu et al. 2020; Lin et al. 2021; Dong et al. 2022; Kou et al. 2022). Among these, E2, Tof5 and Tof12 were associated with domestication (Wang et al. 2016; Lu et al. 2020; Kou et al. 2022) and show sequence diversity among cultivated soybean and its annual relative G. soja. Parallel selection was observed for Tof5 loci in cultivated soybean and its annual wild relatives, both leading to adaptation to high latitudes (Dong et al. 2022). Tof12 is a PRR3 homeolog, loss of Tof12 function had a significant role in adaptation of wild soybean during a phase of initial cultivation and domestication, analysis of the geographic distribution of Tof12 variation suggested the gene may have permitted gradual expansion and improvement at the northern limit of early soybean cultivation (Lu et al. 2020). Tof18 (SOC1a) was reported to be associated with latitude adaption as the Tof18G allele facilitates adaptation to high latitudes, whereas Tof18A facilitates adaptation to low latitudes; however, comparison of sequences of this loci between cultivated soybean and its annual wild relatives suggested it is not a domestication genes (Kou et al. 2022). Thus, G. soja has only limited utility for genetic improvement aimed at the geographic expansion of soybean. By contrast, the wild perennial Glycine spp. are found throughout the tropical and subtropical monsoonal belts of northern and eastern Australia, Papua New Guinea, and the Philippines. Species belonging to the Glycine subgenus are less sensitive to daylength, as suggested by Marshall and Broue (1981). Kenworth et al. (1989) evaluated the daylength sensitivity of 25 accessions of G. tomentella Hayata and one accession each of G. arenaria Tind. and G. tabacina (Labill.) Benth. under various day/night conditions. Accessions from North Queensland in the D4 and T2 isozyme groups were the least sensitive to daylength and produced both flower types under all photoperiods tested, offering a potential source of new germplasm for the improvement of soybean (Kenworth et al. 1989).
Future prospects
Low genetic diversity poses a major challenge to soybean breeding. Wild relative resources can only be used to improve soybean if mapping populations are developed and if alleles associated with beneficial agronomic traits are identified through QTL mapping, association analysis, combined multiple omics approaches, and comparative genomic analysis. The utilization of wild soybean, particularly the perennial Glycine spp., requires further in-depth study of the molecular mechanisms underlying phenotypic traits to better facilitate soybean breeding. Constructing reference sequences and resequencing the genomes of soybean’s wild relatives will allow us to better characterize their genomic features. A coordinated effort is required to generate, analyze, and share data. Soybean breeders will only be able to make use of the available genetic resources if powerful computational resources and bioinformatics tools are developed and if databases for annual and perennial soybean are integrated.
Data availability
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
References
Akpertey A, Singh RJ, Diers BW, Graef GL, Mian MAR, Shannon JG, Scaboo AM, Hudson ME, Thurber CS, Brown PJ, Nelson RL (2018) Genetic introgression from Glycine tomentella to soybean to increase seed yield. Crop Sci 58:1277–1291
Bauer S, Hymowitz T, Noel GR (2007) Soybean cyst nematode resistance derived from Glycine tomentella in amphiploid (G. max × G. tomentella) hybrid lines. Nematropica 37:277–285
Burdon JJ, Marshall DR (1981) Evaluation of Australian native species of Glycine for resistance to soybean rust. Plant Dis 65:44–45
Carter TE, Nelson R, Sneller CH, Cui Z (2004) In soybeans: improvement, production, and uses. In: Boerma HR, Specht JE (eds) Am Soc of Agronomy, Crop Sci Soc of Am, Soil Sci Soc Am J, Madison, WI, vol 16, pp 303–416
Chen P, Yun K, Shao HB, Zhao SJ (2013) Physiological mechanisms for high salt tolerance in wild soybean (Glycine soja) from Yellow River Delta, China: photosynthesis, osmotic regulation, ion flux and antioxidant capacity. PLoS ONE 8:e83227
Coate JE, Luciano AK, Seralathan V, Minchew KJ, Owens TG, Doyle JJ (2012) Anatomical, biochemical and photosynthetic responses to recent allopolyploidy in Glycine dolichocarpa (Fabaceae). Am J Bot 99:55–67
Coate JE, Powell AF, Owens TG, Doyle JJ (2013) Transgressive physiological and transcriptomic responses to light stress in allopoly-ploid Glycine dolichocarpa (Leguminosae). Heredity 110:160–170
Cober ER (2011) Long juvenile soybean flowering responses under very short photoperiods. Crop Sci 51:140–145
Cober ER, Morrison MJ (2010) Regulation of seed yield and agronomic characters by photoperiod sensitivity and growth habit genes in soybean. Theor Appl Genet 120:1005–1012
Cober ER, Voldeng HD (2001) A new soybean maturity and photoperiod-sensitivity locus linked to E1 and T. Crop Sci 41:698–701
Concibido VC, Diers BW, Arelli PR (2004) A decade of QTL mapping for cyst nematode resistance in soybean. Crop Sci 44:1121–1131
Dong LD, Cheng Q, Fang C, Kong LP, Yang H, Hou ZH, Li YL, Nan HY, Zhang YH, Chen QS, Zhang CB, Kou K, Su T, Wang LS, Li SC, Li HY, Lin XY, Tang Y, Zhao XH, Lu SJ, Liu BH, Kong FJ (2022) Parallel selection of distinct Tof5 alleles drove the adaptation of cultivated and wild soybean to high latitudes. Mol Plant 15:308–321
Dwivedi S, Perotti E, Ortiz R (2008) Towards molecular breeding of reproductive traits in cereal crops. Plant Biotechnol J 6:529–559
Gai JY, Zhao TJ (2001) The core ancestors of soybean cultivars in China. J Nanjing Agric Univ 24:20–23
Guan RX, Chen JG, Jiang JH, Liu GY, Liu Y, Tian L, Yu LL, Chang RZ, Qiu LJ (2014) Mapping and validation of a dominant salt tolerance gene in the cultivated soybean (Glycine max) variety Tiefeng 8. Crop J 2:358–365
Guo J, Liu YF, Wang YS, Chen JJ, Li YH, Huang HW, Qiu LJ, Wang Y (2012) Population structure of the wild soybean (Glycine soja) in China: implications from microsatellite analyses. Ann Bot 110:777–785
Harlan JR, de Wet JMJ (1971) Toward a rational classification of cultivated plants. Taxon 20:509–517
Hartman GL (2015) Worldwide importance of soybean pathogens and pests. In: Hartman GL, Rupe JC, Sikora EF, Domier LL, Davis JA, Steffey KL (eds) Compendium of soybean diseases and pests,APS, pp 4–5
Hartman GL, Wang TC, Hymowitz T (1992) Source of resistance to soybean rust in perennial Glycine species. Plant Dis 76:396–399
Hartman GL, Gardner ME, Hymowitz T, Naidoo GC (2000) Evaluation of perennial Glycine species for resistance to soybean fungal pathogens that cause Sclerotinia stem rot and sudden death syndrome. Crop Sci 40:545–549
He SL, Wang YS, Volis S, Li DZ, Yi TS (2012) Genetic diversity and population structure: Implications for conservation of wild soybean (Glycine soja Sieb. et Zucc) based on nuclear and chloroplast microsatellite variation. Int J Mol Sci 13:12608–12628
Herman TK, Han J, Singh RJ, Domier LL, Hartman GL (2020) Evaluation of wild perennial Glycine species for resistance tosoybean cyst nematode and soybean rust. Plant Breed 139:923–931
Hymowitz T (1970) On the domestication of the soybean. Econ Bot 24:408–421
Hymowitz T (1995) Evaluation of wild perennial Glycine species and crosses for resistance to Phakospora. In: Sinclair et al. (ed) Proceedings of the Soybean Rust Workshop. NSRL, Urbana, pp 33–37
Hyten DL, Song QJ, Zhu YL, Choi IY, Nelson RL, Costa JM, Specht JE, Shoemaker RC, Cregan PB (2006) Impacts of genetic bottlenecks on soybean genome diversity. Proc Natl Acad Sci USA 103:16666–16671
Hyten DL, Choi IY, Song QJ, Shoemaker RC, Nelson RL, Costa JM, Specht JE, Cregan PB (2007) Highly variable patterns of linkage disequilibrium in multiple soybean populations. Genetics 175:1937–1944
Ilut DC, Coate JE, Luciano AK, Owens TG, May GD, Farmer A, Doyle JJ (2012) A comparative transcriptomic study of an allotetraploid and its diploid progenitors illustrates the unique advantages and challenges of RNA-seq in plant species. Am J Bot 99:383–396
Innes RW, Ameline-Torregrosa C, Ashfield T, Cannon E, Cannon S, Chacko B, Chen N, Couloux A, Dalwani A, Denny R, Deshpande S, Egan AN, Glover N, Hans CS, Howell S, Ilut DC, Jackson S, Lai HS, Mammadov JA, Martin S, Metcalf M, Nguyen A, O’Bleness M, Pfeil BE, Podicheti R, Ratnaparkhe MB, Samain S, Sanders I, Ségurens B, Sévignac M, Sherman-Broyles S, Thareau V, Tucker DM, Walling J, Wawrzynski A, Yi J, Doyle JJ, Geffroy V, Roe BA, Maroof MAS, Young ND (2008) Differential accumulation of retroelements and diversification of NB-LRR disease resistance genes in duplicated regions following polyploidy in the ancestor of soybean. Plant Physiol 148:1740–1759
Ji W, Zhu YM, Li Y, Yang L, Zhao XW, Cai H, Bai X (2010) Over-expression of a glutathione S-transferase gene, GsGST, from wild soybean (Glycine soja) enhances drought and salt tolerance in transgenic tobacco. Biotechnol Lett 32:1173–1179
Kao WY, Tsai TT (1998) Tropic leaf movements, photosynthetic gas exchange, leaf δ13C and chlorophyll a fluorescence of three soybean species in response to water availability. Plant Cell Environ 21:1055–1062
Kao WY, Tsai TT, Shih CN (2003) Photosynthetic gas exchange and chlorophyll a fluorescence of three wild soybean species in response to NaCl treatments. Photosynthetica 41:415–419
Kao WY, Tsai TT, Tsai HC, Shih CN (2006) Response of three Glycine species to salt stress. Environ Exp Bot 56:120–125
Katerji N, Hoorn JW, Hamdy A, Mastrorilli M (2003) Salinity effect on crop development and yield, analysis of salt tolerance according to several classification methods. Agric Water Manage 62:37–66
Kenworthy WJ (1989) Potential genetic contributions of wild relatives to soybean improvements. In: Pascale AJ (ed) World Research Conference. Assoc. Soja, Buenos Aires, Argentina, pp 883–888
Kenworthy WJ, Brown A, Thibou GA (1989) Variation in flowering response to photoperiod in perennial glycine species. Crop Sci 29:678–682
Kim M, Diers BW (2013) Fine mapping of the SCN resistance QTL cqSCN-006 and cqSCN-007 from Glycine soja PI 468916. Crop Sci 53:775–785
Kim MY, Lee S, Van K, Kim TH, Jeong SC, Choi IY, Kim DS, Lee YS, Park D, Ma JX, Kim WY, Kim BC, Park S, Lee KA, Kim DH, Kim KH, Shin JH, Jang YE, Kim KD, Liu WX, Chaisan T, Kang YJ, Lee YH, Kim KH, Moon JK, Schmutz J, Jackson SA, Bhak J, Lee SH (2010) Whole-genome sequencing and intensive analysis of the undomesticated soybean (Glycine soja Sieb. and Zucc.) genome. Proc Natl Acad Sci USA 107:22032–22037
Kim M, Hyten DL, Niblack TL, Diers BW (2011) Stacking resistance alleles from wild and domestic soybean sources improves soybean cyst nematode resistance. Crop Sci 51:934–943
Kim H, Kim ST, Ryu J, Kang BC, Kim JS, Kim SG (2017) CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat Commun 8:14406
Kofsky J, Zhang HY, Song BH (2018) The untapped genetic reservoir: the past, current, and future applications of the wild soybean (Glycine soja). Front Plant Sci 9:949
Kong FJ, Liu BH, Xia ZJ, Sato S, Kim BM, Watanabe S, Yamada T, Tabata S, Kanazawa A, Harada K, Abe J (2010) Two coordinately regulated homologs of FLOWERING LOCUS T are involved in the control of photoperiodic flowering in soybean. Plant Physiol 154:1220–1231
Kou K, Yang H, Li HY, Fang C, Chen LY, Yue L, Nan HY, Kong LP, Li XM, Wang F, Wang JH, Du HP, Yang ZY, Bi YD, Lai YC, Dong LD, Cheng Q, Su T, Wang LS, Li SC, Hou ZH, Lu SJ, Zhang YH, Che ZJ, Yu DY, Zhao XH, Liu BH, Kong FJ (2022) A functionally divergent SOC1 homolog improves soybean yield and latitudinal adaptation. Curr Biol 32:1–15
Lam HM, Xu X, Liu X, Chen WB, Yang GH, Wong FL, Li MW, He WM, Qin N, Wang B, Li J, Jian M, Wang J, Shao GH, Wang J, Sun SSM, Zhang GY (2010) Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat Genet 42:1053–1059
Le DT, Nishiyama R, Watanabe Y, Tanaka M, Seki M, Ham LH, Yamaguchi-Shinozaki K, Shinozaki K, Tran LSP (2012) Differential gene expression in soybean leaf tissues at late developmental stages under drought stress revealed by genome-wide transcriptome analysis. PLoS ONE 7:e49522
Lee JD, Shannon JG, Vuong TD, Nguyen HT (2009) Inheritance of salt tolerance in wild soybean (Glycine soja Sieb. and Zucc.) accession PI483463. J Hered 100:798–801
Lee JS, Yoo MH, Jung JK, Bilyeu KD, Lee JD, Kang S (2015) Detection of novel QTLs for foxglove aphid resistance in soybean. Theor Appl Genet 128:1481–1488
Lenis JM, Ellersieck M, Blevins DG, Sleper DA, Nguyen HT, Dunn D, Lee JD, Shannon JG (2011) Differences in ion accumulation and salt tolerance among Glycine accessions. J Agron Crop Sci 197:302–310
Li YH, Guan RX, Liu ZX, Ma YS, Wang LX, Li LH, Lin FY, Luan WJ, Chen PY, Yan Z, Guan Y, Zhu L, Ning XC, Smulders MJ, Li W, Piao RH, Cui YH, Yu ZM, Guan M, Chang RZ, Hou AF, Shi AN, Zhang B, Zhu SL, Qiu LJ (2008) Genetic structure and diversity of cultivated soybean (Glycine max (L.) Merr.) landraces in China. Theor Appl Genet 117:857–871
Li X, Huang L, Lu JH, Cheng YH, You QB, Wang LJ, Song XJ, Zhou XN, Jiao YQ (2018) Large-scale investigation of soybean gene functions by overexpressing a full-length soybean cDNA library in Arabidopsis. Front Plant Sci 9:631
Li YH, Zhou GY, Ma JX, Jiang WK, Jin LG, Zhang ZH, Guo Y, Zhang JB, Sui Y, Zheng LT, Zhang SS, Zuo QY, Shi XH, Li YF, Zhang WK, Hu YY, Kong GY, Hong HL, Tan B, Song J, Liu ZX, Wang YS, Ruan H, Yeung CKL, Liu J, Wang HL, Zhang LJ, Guan RX, Wang KJ, Li WB, Chen SY, Chang RZ, Jiang Z, Jackson SA, Li RQ, Qiu LJ (2014) De novo assembly of soybean wild relatives for pan-genome analysis of diversity and agronomic traits. Nat Biotechnol 32:1045–1052
Lim SM, Hymowitz T (1987) Reactions of perennial wild species of genus Glycine to Septoria glycines. Plant Dis 71:891–893
Lin XY, Liu BH, Weller JL, Abe J, Kong FJ (2021) Molecular mechanisms for the photoperiodic regulation of flowering in soybean. J Integr Plant Biol 63:981–994
Liu B, Kanazawa A, Matsumura H, Takahashi R, Harada K, Abe J (2008) Genetic redundancy in soybean photoresponses associated with duplication of the phytochrome A gene. Genetics 180:995–1007
Liu Q, Chang SY, Hartman GL, Domier LL (2018) Assembly and annotation of a draft genome sequence for Glycine latifolia, a perennial wild relative of soybean. Plant J 95:71–85
Liu YC, Du HL, Li PC, Shen YT, Peng H, Liu SL, Zhou GA, Zhang HK, Liu Z, Shi M, Huang XH, Li Y, Zhang M, Wang Z, Zhu BG, Han B, Liang CZ, Tian ZX (2020) Pan-genome of wild and cultivated soybeans. Cell 182:162–176
Lu SJ, Zhao XH, Hu YL, Liu SL, Nan HY, Li XM, Fang C, Cao D, Shi XY, Kong LP, Su T, Zhang FG, Li SC, Wang Z, Yuan XH, Cober ER, Weller JL, Liu BH, Hou XL, Tian ZX, Kong FJ (2017) Natural variation at the soybean J locus improves adaptation to the tropics and enhances yield. Nat Genet 49:773–779
Lu SJ, Dong LD, Fang C, Liu SL, Kong LP, Cheng Q, Chen LY, Su T, Nan HY, Zhang D, Zhang L, Wang ZJ, Yang YQ, Yu DY, Liu XL, Yang QY, Lin XY, Tang Y, Zhao XH, Yang XQ, Tian CE, Xie QG, Li X, Yuan XH, Tian ZX, Liu BH, Weller JL, Kong FJ (2020) Stepwise selection on homeologous PRR genes controlling flowering and maturity during soybean domestication. Nat Genet 52:428–436
Marshal DR, Broue P (1981) The wild relatives of crop plants, indigenous to Australia and their use in plant breeding. J Aust Inst Agric Sci 47:149–154
Melito S, Heuberger AL, Cook D, Diers BW, MacGuidwin AE, Bent AF (2010) A nematode demographics assay in transgenic roots reveals no significant impacts of the Rhg1locus LRR-Kinase on soybean cyst nematode resistance. BMC Plant Biol 10:104
Mignucci JS, Chamberlain DW (1978) Interactions of Microsphaera diffusa with soybeans and other legumes. Phytopathology 68:169–173
Morrell PL, Toleno DM, Lundy KE, Clegg MT (2005) Low levels of linkage disequilibrium in wild barley (Hordeum vulgare ssp. spontaneum) despite high rates of self-fertilization. Proc Natl Acad Sci USA 102:2442–2447
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59:651–681
Niblack TL, Riggs RD (2015) Soybean Cyst Nematode. In: Hartman GL, Rupe JC, Sikora EF, Domier LL, Davis JA, Steffey KL (eds) Compendium of soybean diseases and pests, APS, pp 100–104
Ning WF, Zhai H, Liang YuJQ, S, Yang X, Xing XY, Huo JL, Pang T, Yang YL, Bai X, (2017) Overexpression of Glycine soja WRKY20 enhances drought tolerance and improves plant yields under drought stress in transgenic soybean. Mol Breeding 37:19
Ohigashi K, Mizuguti A, Nakatani K, Matsuo K (2019) Modeling the flowering sensitivity of five accessions of wild soybean (Glycine soja) to temperature and photoperiod, and its latitudinal cline. Breeding Sci 69:84–93
Pantalone VR, Kenworthy WJ, Slaughter LH, James BR (1997) Chloride tolerance in soybean and perennial Glycine accessions. Euphytica 97:235–239
Phang TH, Shao GH, Lam HM (2008) Salt tolerance in soybean. J Integr Plant Biol 50:1196–1212
Qi XP, Li MW, Xie M, Liu X, Ni M, Shao GH, Song C, Yim AKY, Tao Y, Wong FL, Isobe S, Wong CF, Wong KS, Xu CY, Li CQ, Wang Y, Guan R, Sun FM, Fan GY, Xiao ZX, Zhou F, Phang TH, Liu X, Tong SW, Chan TF, Yiu SM, Tabata S, Wang J, Xu X, Lam HM (2014) Identification of a novel salt tolerance gene in wild soybean by whole-genome sequencing. Nat Commun 5:4340
Ray DK, Mueller ND, West PC, Foley JA (2013) Yield trends are insufficient to double global crop production by 2050. PLoS ONE 8:e66428
Riggs RD, Schmitt DP (1988) Complete characterization of the race scheme for Heterodera glycines. J Nematol 20:392–395
Schmid KJ, Ramos-Onsins S, Ringys-Beckstein H, Weisshaar B, Mitchell-Olds T (2005) A multilocus sequence survey in Arabidopsis thaliana reveals a genome-wide departure from a neutral model of DNA sequence polymorphism. Genetics 169:1601–1615
Schmutz J, Cannon SB, Schlueter J, Ma JX, Mitros T, Nelson W, Hyten DL, Song QJ, Thelen JJ, Cheng JL, Xu D, Hellsten U, May GD, Yu Y, Sakurai T, Umezawa T, Bhattacharyya MK, Sandhu D, Valliyodan B, Lindquist E, Peto M, Grant D, Shu SQ, Goodstein D, Barry K, Futrell-Griggs M, Abernathy B, Du JC, Tian ZX, Zhu LC, Gill N, Joshi T, Libault M, Sethuraman A, Zhang XC, Shinozaki K, Nguyen HT, Wing RA, Cregan P, Specht J, Grimwood J, Rokhsar D, Stacey G, Shoemaker RC, Jackson SA (2010) Genome sequence of the palaeopolyploid soybean. Nature 463:178–183
Sedivy EJ, Wu FQ, Hanzawa Y (2017) Soybean domestication: the origin, genetic architecture and molecular bases. New Phytol 214:539–553
Sherman-Broyles S, Bombarely A, Powell AF, Doyle JL, Egan AN, Coate JE, Doyle JJ (2014) The wild side of a major crop: soybean’s perennial cousins from Down Under. Am J Bot 101:1651–1665
Singh RJ, Hymowitz T (2011) Soybean genetic resourcees and crop improvement. Genome 42:605–616
Singh RJ, Nelson RL, Chung G (2007) Genetic resources, chromosome engineering, and crop improvement series. In: Ram JS (ed) Oilseed crops, volume 4, CRC Press (Taylor & Francis Group), Boca Raton
Singh RJ (2017) Botany and cytogenetics of soybean. In: Nguyen MK, Bhattacharyya HT (eds) The Soybean Genome
Song B, Oehrle NW, Liu SS, Krishnan HB (2016) Characterization of seed storage proteins of several perennial Glycine species. J Agric Food Chem 64:8499–8508
Tanksley SD, McCouch SR (1997) Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277:1063–1066
Tian JG, Wang CL, Xia JL, Wu LS, Xu GH, Wu WH, Li D, Qin WC, Han X, Chen QY, Jin WW, Tian F (2019) Teosinte ligule allele narrows plant architecture and enhances high-density maize yields. Science 365:658–664
Tilman D, Balzer C, Hill J, Befort BL (2011) Global food demand and the sustainable intensification of agriculture. Proc Natl Acad Sci USA 108:20260–20264
Wang Y, Gu YZ, Gao HH, Qiu LJ, Chang RZ, Chen SY, He CY (2016) Molecular and geographic evolutionary support for the essential role of GIGANTEAa in soybean domestication of flowering time. BMC Evol Biol 16:79
Wang FF, Nan HY, Chen LY, Fang C, Zhang HY, Su T, Li SC, Cheng Q, Dong LD, Liu BH, Kong FJ, Lu SJ (2019) A new dominant locus, E11, controls early flowering time and maturity in soybean. Mol Breeding 39:70
Watanabe S, Hideshima R, Xia ZJ, Tsubokura Y, Sato S, Nakamoto Y, Yamanaka N, Takahashi R, Ishimoto M, Anai T, Tabata S, Harada K (2009) Map-based cloning of the gene associated with the soybean maturity Locus E3. Genetics 182:1251–1262
Watanabe S, Xia ZJ, Hideshima R, Tsubokura Y, Sato S, Yamanaka N, Takahashi R, Anai T, Tabata S, Kitamura K, Harada K (2011) A map-based cloning strategy employing a residual heterozygous line reveals that the GIGANTEA gene is involved in soybean maturity and flowering. Genetics 188:395–407
Wawrzynski A, Ashfield T, Chen NWG, Mammadov J, Nguyen A, Podicheti R, Cannon SB, Thareau V, Ameline-Torregrosa C, Cannon E, Chacko B, Couloux A, Dalwani A, Denny R, Deshpande S, Egan AN, Glover N, Howell S, Ilut D, Lai HS, del Campo SM, Metcalf M, O’Bleness M, Pfeil BE, Ratnaparkhe MB, Samain S, Sanders I, Ségurens B, Sévignac M, Sherman-Broyles S, Tucker DM, Yi J, Doyle JJ, Geffroy V, Roe BA, Maroof MAS, Young ND, Innes RW (2008) Replication of nonautonomous retroelements in soybean appears to be both recent and common. Plant Physiol 148:1760–1771
Wen L, Yuan C, Herman TK, Hartman GL (2017) Accessions of perennial Glycine species with resistance to multiple types of soybean cyst nematode (Heterodera glycines). Plant Dis 101:1201–1206
Wright SI, Bi IV, Schroeder SG, Yamasaki M, Doebley JF, McMullen MD, Gaut BS (2005) The effects of artificial selection on the maize genome. Science 308:1310–1314
Xia ZJ, Watanabe S, Yamada T, Tsubokura Y, Nakashima H, Zhai H, Anai T, Sato S, Yamazaki T, Lü SX, Wu HY, Tabata S, Harada K (2012) Positional cloning and characterization reveal the molecular basis for soybean maturity locus E1 that regulates photoperiodic flowering. Proc Natl Acad Sci USA 109:E2155-2164
Xie M, Chung CYL, Li MW, Wong FL, Wang X, Liu AL, Wang ZL, Leung AKY, Wong TH, Tong SW, Xiao ZX, Fan KJ, Ng MS, Qi XP, Yang LF, Deng TQ, He LJ, Chen L, Fu AS, Ding Q, He JX, Chung G, Isobe S, Tanabata T, Valliyodan B, Nguyen HT, Cannon SB, Foyer CH, Chan TF, Lam HM (2019) A reference-grade wild soybean genome. Nat Commun 10:1216
Xue ZC, Zhao SJ, Gao HY, Sun S (2014) The salt resistance of wild soybean (Glycine soja Sieb. et Zucc. ZYD 03262) under NaCl stress is mainly determined by Na+ distribution in the plant. Acta Physiol Plant 36:61–70
Zhai H, Lü SX, Liang S, Wu HY, Zhang XZ, Liu BH, Kong FJ, Yuan XH, Li J, Xia ZJ (2014) GmFT4, a homolog of FLOWERING LOCUS T, is positively regulated by E1 and functions as a flowering repressor in soybean. PLoS ONE 9:e89030
Zhang FN, Batley J (2020) Exploring the application of wild species for crop improvement in a changing climate. Curr Opin Plant Biol 56:218–222
Zhang GR, Gu CH, Wang DC (2013) Mapping and validation of a gene for soybean aphid resistance in PI 567537. Mol Breeding 32:131–138
Zhang HY, Li CY, Davis EL, Wang JS, Griffin JD, Kofsky J, Song BH (2016) Genome-wide association study of resistance to soybean cyst nematode (Heterodera glycines) HG Type 2.5.7 in wild soybean (Glycine soja). Front Plant Sci 7:1214
Zhang SC, Zhang ZN, Bales C, Gu CH, DiFonzo C, Li M, Song QJ, Cregan P, Yang ZY, Wang DC (2017a) Mapping novel aphid resistance QTL from wild soybean, Glycine soja 85–32. Theor Appl Genet 130:1941–1952
Zhang SC, Zhang ZN, Wen ZX, Gu CH, An YC, Bales C, DiFonzo C, Song QJ, Wang DC (2017b) Fine mapping of the soybean aphid-resistance genes Rag6 and Rag3c from Glycine soja 85–32. Theor Appl Genet 130:2601–2615
Zhang DY, Kumar M, Xu L, Wan Q, Huang YH, Xu ZL, He XL, Ma JB, Pandey GK, Shao HB (2017c) Genome-wide identification of Major Intrinsic Proteins in Glycine soja and characterization of GmTIP2;1 function under salt and water stress. Sci Rep 7:4106
Zhang HF, Yasmin F, Song BH (2019) Neglected treasures in the wild—legume wild relatives in food security and human health. Curr Opin Plant Biol 49:17–26
Funding
This work was supported by the National Key Research and Development Program (grant no. 2021YFF1001203), the Taishan Scholars Program of Shandong Province (tsqn201812036), the Agricultural Variety Improvement Project of Shandong Province (2019LZGC004), and Program for Scientific Research Innovation Team of Young Scholar in Colleges and Universities of Shandong Province, China (2020KJF008).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
No potential conflict of interest was disclosed.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhuang, Y., Li, X., Hu, J. et al. Expanding the gene pool for soybean improvement with its wild relatives. aBIOTECH 3, 115–125 (2022). https://doi.org/10.1007/s42994-022-00072-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42994-022-00072-7