Abstract
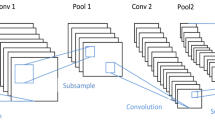
The percentage of people affected by skin cancer has been rising in recent years. Melanoma is identified as the most dangerous and life-threatening among the three types of skin cancer since it causes more deaths than the other two over time. According to the expertise, if the melanoma case is discovered at an early stage, the death rate may be decreased. Due to the skin lesion’s complexity, it is challenging to identify melanoma at an early stage. In this work, an automated assistant system is suggested to help doctors in identifying melanoma effectively at an early stage. Because pixel intensity values include distinctive and useful features in an image, hence the pixel intensity value estimation (IVE) model is embedded with a transfer learning network for efficient Melanoma detection. Four popular transfer learning models have been analyzed to derive the best-performed model in Melanoma detection (MD). Finally, data sensitivity is analyzed on the best model. The experiment shows that overall the best performance in Recall (98.8%), F1-score (99.0%), Accuracy (99.18%), and AUC-ROC curve (97.8%) is achieved by the VGG-16 transfer learning model for 4056 data; we denoted the model as IVE-MDNet model. In the model, the network consists of 13 convolutional layers and five max-pooling layers and the learning weights are obtained using the ‘ImageNet’ dataset. A new sub-layer model is formed, which is combined with the pre-trained network to design the proposed transfer learning approach. Before feeding the image to the model, the artifacts were removed using a pre-processing technique which uses a series of precise procedures.






Similar content being viewed by others
Data Availability
The authors declared that all of the material is owned by the authors and/or no permissions are required.
References
Adegun A, Viriri S. An enhanced deep learning framework for skin lesions segmentation. In: International conference on computational collective intelligence. Berlin: Springer; 2019. p. 414–25.
Agarwal S, Rattani A, Chowdary CR. A comparative study on handcrafted features v/s deep features for open-set fingerprint liveness detection. Pattern Recogn Lett. 2021;147:34–40.
Agilandeeswari L, Sagar MT, Keerthana N. Skin lesion detection using texture based segmentation and classification by convolutional neural networks (CNN). Art Int J Innov Technol Explor Eng. 2019;9(2):2117–21120.
American Cancer Society. Key statistics for melanoma skin cancer. https://www.cancer.org/cancer/melanoma-skin-cancer/about, 2022.
Argenziano G, Fabbrocini G, Carli P, De Giorgi V, Sammarco E, Delfino M. Epiluminescence microscopy for the diagnosis of doubtful melanocytic skin lesions: comparison of the ABCD rule of dermatoscopy and a new 7-point checklist based on pattern analysis. Arch Dermatol. 1998;134(12):1563–70.
Ashafuddula NI, Islam R. Melanoma skin cancer and nevus mole classification using intensity value estimation with convolutional neural network. Comput Sci. 2023;24(3).
Baldi S, Michailidis I, Ntampasi V, Kosmatopoulos EB, Papamichail I, Papageorgiou M. Simulation-based synthesis for approximately optimal urban traffic light management. In: 2015 American control conference (ACC). New York: IEEE; 2015. p. 868–873.
Carli P, Quercioli E, Sestini S, Stante M, Ricci L, Brunasso G, De Giorgi V. Pattern analysis, not simplified algorithms, is the most reliable method for teaching dermoscopy for melanoma diagnosis to residents in dermatology. Br J Dermatol. 2003;148(5):981–4.
Codella N, Cai J, Abedini M, Garnavi R, Halpern A, Smith JR. Deep learning, sparse coding, and SVM for melanoma recognition in dermoscopy images. In: International workshop on machine learning in medical imaging. Berlin: Springer; 2015. p. 118–26.
Eltayef K, Li Y, Liu X. Detection of melanoma skin cancer in dermoscopy images. In: Journal of physics: conference series, vol. 787. IOP Publishing; 2017. p. 012034.
Fatima R, Zafar Ali Khan M, Govardhan A, Dhruve KD. Computer aided multi-parameter extraction system to aid early detection of skin cancer melanoma. Int J Comput Sci Netw Secur. 2012;12(10):74–86.
Garg R, Maheshwari S, Shukla A. Decision support system for detection and classification of skin cancer using CNN. arXiv preprint arXiv:1912.03798. 2019.
Gerges F, Shih FY. A convolutional deep neural network approach for skin cancer detection using skin lesion images. Int J Electr Comput Eng. 2021;15(8):475–8.
Giotis I, Molders N, Land S, Biehl M, Jonkman MF, Petkov N. Med-node: a computer-assisted melanoma diagnosis system using non-dermoscopic images. Expert Syst Appl. 2015;42(19):6578–85.
Gogul I, Sathiesh KV. Flower species recognition system using convolution neural networks and transfer learning. In: 2017 fourth international conference on signal processing, communication and networking (ICSCN). New York: IEEE; 2017. p. 1–6.
Yanming Guo Yu, Liu AO, Lao S, Song W, Lew MS. Deep learning for visual understanding: a review. Neurocomputing. 2016;187:27–48.
He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition. 2016. p. 770–8.
Hosny KM, Kassem MA, Foaud MM. Classification of skin lesions using transfer learning and augmentation with Alex-net. PLoS ONE. 2019;14(5):e0217293.
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ. Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition. 2017. p. 4700–8.
Islam R, Imran S, Ashikuzzaman M, Khan MMA. Detection and classification of brain tumor based on multilevel segmentation with convolutional neural network. J Biomed Sci Eng. 2020;13(4):45–53.
Kassani SH, Kassani PH. A comparative study of deep learning architectures on melanoma detection. Tissue Cell. 2019;58:76–83.
Kaur T, Gandhi TK. Deep convolutional neural networks with transfer learning for automated brain image classification. Mach Vis Appl. 2020;31(3):1–16.
Khan MQ, Hussain A, Rehman SU, Khan U, Maqsood M, Mehmood K, Khan MA. Classification of melanoma and nevus in digital images for diagnosis of skin cancer. IEEE Access. 2019;7:90132–44.
Lakshminarayanan AR, Bhuvaneshwari R, Bhuvaneshwari S, Parthasarathy S, Jeganathan S, Sagayam KM. Skin cancer prediction using machine learning algorithms. In: Artificial intelligence and technologies. Berlin: Springer; 2022. p. 303–10.
LeCun Y, Boser B, Denker JS, Henderson D, Howard RE, Hubbard W, Jackel LD. Backpropagation applied to handwritten zip code recognition. Neural Comput. 1989;1(4):541–51.
Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, Van Der Laak JA, Van Ginneken B, Sánchez CI. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88.
Maiti A, Chatterjee B. Improving detection of melanoma and naevus with deep neural networks. Multimed. Tools Appl. 2019:1–20.
Manzo M, Pellino S. Bucket of deep transfer learning features and classification models for melanoma detection. J Imaging. 2020;6(12):129.
Mishra NK, Celebi ME. An overview of melanoma detection in dermoscopy images using image processing and machine learning. arXiv preprint arXiv:1601.07843, 2016.
Mukherjee S, Adhikari A, Roy M. Malignant melanoma detection using multi layer preceptron with visually imperceptible features and PCA components from med-node dataset. Int J Med Eng Inform. 2020;12(2):151–68.
Nasr-Esfahani E, Samavi S, Karimi N, Soroushmehr SM, Jafari MH, Ward K, Najarian K. Melanoma detection by analysis of clinical images using convolutional neural network. In: 2016 38th annual international conference of the IEEE engineering in medicine and biology society (EMBC). New York: IEEE; 2016. p. 1373–6.
Peram MR, Jalalpure S, Kumbar V, Patil S, Joshi S, Bhat K, Diwan P. Factorial design based curcumin ethosomal nanocarriers for the skin cancer delivery: in vitro evaluation. J Liposome Res. 2019;29(3):291–311.
Pillay V, Hirasen D, Viriri S, Gwetu M.. Melanoma skin cancer classification using transfer learning. In: International conference on computational collective intelligence. Berlin: Springer; 2020. p. 287–97.
Pillay V, Hirasen D, Viriri S, Gwetu M. Melanoma skin cancer classification using transfer learning. In: International conference on computational collective intelligence. Berlin: Springer; 2020. p. 287–97.
Refianti R, Mutiara AB, Priyandini RP. Classification of melanoma skin cancer using convolutional neural network. Int J Adv Comput Sci Appl. 2019;10(3):409–17.
Shorten C, Khoshgoftaar TM. A survey on image data augmentation for deep learning. J Big Data. 2019;6(1):1–48.
Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556, 2014.
Sultana NN, Mandal B, Puhan NB. Deep residual network with regularised fisher framework for detection of melanoma. IET Comput Vis. 2018;12(8):1096–104.
Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z. Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE conference on computer vision and pattern recognition. 2016. p. 2818–26.
Tan C, Sun F, Kong T, Zhang W, Yang C, Liu C. A survey on deep transfer learning. In: International conference on artificial neural networks. Berlin: Springer; 2018. p. 270–9.
Torrey L, Shavlik J. Transfer learning. In: Handbook of research on machine learning applications and trends: algorithms, methods, and techniques. IGI global; 2010. p. 242–64.
Vestergaard ME, Macaskill PHPM, Holt PE, Menzies SW. Dermoscopy compared with naked eye examination for the diagnosis of primary melanoma: a meta-analysis of studies performed in a clinical setting. Br J Dermatol. 2008;159(3):669–76.
Vijayalakshmi MM. Melanoma skin cancer detection using image processing and machine learning. Int J Trend Sci Res Dev. 2019;3(4):780–4.
Yadav AK, Roy R, Kumar AP, et al. Survey on content-based image retrieval and texture analysis with applications. Int J Signal Process Image Process Pattern Recogn. 2014;7(6):41–50.
Yadav SS, Jadhav SM. Deep convolutional neural network based medical image classification for disease diagnosis. J Big Data. 2019;6(1):1–18.
Zaman K, Maghdid SS. Medical images classification for skin cancer using convolutional neural network algorithms. Adv Mech. 2021;9(3):526–41.
Zheng C, Sun D-W. Image segmentation techniques. In: Computer vision technology for food quality evaluation. Amsterdam: Elsevier; 2008. p. 37–56.
Zunair H, Hamza AB. Melanoma detection using adversarial training and deep transfer learning. Phys Med Biol. 2020;65(13):135005.
Acknowledgements
The authors wish to thank the Department of Computer Science and Engineering of Dhaka University of Engineering & Technology, Gazipur, for providing research support.
Funding
The authors confirm that the submitted work has not received any funding from any organization.
Author information
Authors and Affiliations
Contributions
Mr. Md. Ashafuddula is currently doing his research work in Computer Science and Engineering department under the supervision of Professor Dr. Rafiqul Islam. Professor Dr. Rafiqul Islam has analyzed the study and planned the research experiment. Both the authors have directly participated in the execution of this work, resulting in and writing the paper equally.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Ethical Approval
This research did not contain any studies involving animal or human participants, nor did it take place on any private or protected areas. No specific permissions were required for corresponding locations.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ashafuddula, N.I.M., Islam, R. IVE-MDNet: Intensity Value Estimation Model Combined with a Transfer Learning Approach for Melanoma Skin Cancer Diagnosis. SN COMPUT. SCI. 5, 435 (2024). https://doi.org/10.1007/s42979-024-02800-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42979-024-02800-w