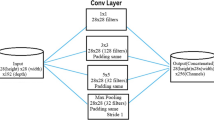
Abstract
Agricultural sector, in association with its allied sectors, plays pivotal role in the progress of a country. In spite of notable contribution, Agricultural sector suffers from several challenges. Crop loss due to diseases is considered as one of the major challenges. Rice is one of the major cereal crops in India. Its production is highly impacted due to crop diseases. Early and accurate detection of rice crop leaf diseases is essential for aforementioned issue. Over the years, conventional methods of crop leaf disease detection are used but with its own limitations. Recent technological advancement in computer vision, deep learning has created new pathways in agricultural sector. Deep learning models require huge data which is quite a great challenge. In such case, building up of a robust deep learning model could work with limited and unbalanced dataset, with good generalization performances. In this study, weighted deep ensemble learning approach is used for performance improvement of rice crop leaf disease classification task. The ensemble method ConvDepthTransEnsembleNet is proposed. To show the effectiveness of proposed work, experiments are conducted on diverse datasets. ConvDepthTransEnsembleNet is a lightweight model which has achieved the accuracy of 96.88% on limited and unbalanced dataset. The experimental results show that proposed model outperforms the individual classifiers based on conventional methods and transfer learning approach in terms of significant reduction in parameters and improved upon generalization performance. The proposed model is highly useful for implementing deep learning models with resources constrained devices.





Similar content being viewed by others
Data Availability Statement
Data will be made available on request.
References
Biswaranjan A, Ahona G, Sucheta P, Yu-Chen H. Automated plant recognition system with geographical position selection for medicinal plants. Adv Multimed. 2023. https://doi.org/10.1155/2023/3974346.
Liang W, Zhang Z, Zhang G, Cao H. Rice blast disease recognition using a deep convolutional neural network. Sci Rep. 2019;9:2869. https://doi.org/10.1038/s41598-019-38966-0.
Dubey SR, Jalal AS. Adapted approach for fruit disease identification using images. In: Information Resources Management Association (eds) Image processing: concepts, methodologies, tools, and applications. Hershey: IGI Global; 2013. pp. 1395–9
Andrew J, Eunice J, Popescu DE, Chowdary MK, Hemanth J. Deep learning-based leaf disease detection in crops using images for agricultural applications. Agronomy. 2022;12(10):2395. https://doi.org/10.3390/agronomy12102395.
Lu Y, Yi S, Zeng N, Liu Y, Zhang Y. Identification of rice diseases using deep convolutional neural networks. Neurocomputing. 2017;267:378–84.
Picon A, Alvarez-Gila A, Seitz M, Ortiz-Barredo A, Echazarra J, Johannes A. Deep convolutional neural networks for mobile capture device-based crop disease classifcation in the wild. Comput Electron Agric. 2019;161:280–90.
Sandler M, Howard A, Zhu M, Zhmoginov A, Chen LC. MobileNetV2: inverted residuals and linear bottlenecks. In: Proceeding of the IEEE conference on computer vision and pattern recognition (CVPR), Salt Lake City, UT, pp. 4510–4520. 2018.
Ma N, Zhang X, Zheng H-T, Sun J. Shufflenet v2: Practical guidelines for efficient CNN architecture design. In: Proceedings of the European conference on computer vision (ECCV), pp 116–131. 2018.
Tan M, Le Q. Efficientnet: rethinking model scaling for convolutional neural networks. In: International conference on machine learning, PMLR, pp. 6105–6114. 2019.
Han K, Wang Y, Tian Q, Guo J, Xu C, Xu C. Ghostnet: more features from cheap operations. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 1580–1589. 2020.
Thakur PS, Sheorey T, Ojha A. VGG-ICNN: a Lightweight CNN model for crop disease identification. Multimed Tools Appl. 2023;82:497–520. https://doi.org/10.1007/s11042-022-13144-z.
Padol PB, Yadav AA. SVM classifier based grape leaf disease detection. In: Conference on Advances in Signal Processing (CASP), Pune, pp. 175–179. 2016. Doi: https://doi.org/10.1109/CASP.2016.7746160
Nawaz MA, et al. Plant disease detection using internet of thing (IoT). Int J Adv Comput Sci Appl. 2020;11:1.
Prajapati HB, Shah JP, Dabhi VK. Detection and classification of rice plant diseases. Intell Decis Technol. 2017;11:357–73. https://doi.org/10.3233/IDT-170301.
Lamba S, Vinay K, Anupam B, Shalli R, Syed HA. A novel hybrid severity prediction model for blast paddy disease using machine learning. Sustainability. 2023;15(2):1502. https://doi.org/10.3390/su15021502.
Sethy PK, Barpanda NK, Rath AK, Behera SK. Deep feature based rice leaf disease identification using support vector machine. Comput Electron Agric. 2020;175: 105527. https://doi.org/10.1016/j.compag.2020.105527.
Bouguettaya A, Zarzour H, Kechida A, et al. A survey on deep learning-based identification of plant and crop diseases from UAV-based aerial images. Clust Comput. 2023;26:1297–317. https://doi.org/10.1007/s10586-022-03627-x.
Pinki FT et al (2017) Content based paddy leaf disease recognition and remedy prediction using support vector machine. In: 20th International Conference of Computer and Information Technology (ICCIT), pp. 1–5. 2017. doi: https://doi.org/10.1109/ICCITECHN.2017.8281764.
Geetharamani G, Arun PJ. Identification of plant leaf diseases using a nine-layer deep convolutional neural network. Comput Electr Eng. 2019;76:323–38. https://doi.org/10.1016/j.compeleceng.2019.04.011.
Upadhyay SK, Kumar A. A novel approach for rice plant diseases classification with deep convolutional neural network. Int J Inf Tecnol. 2022;14:185–99. https://doi.org/10.1007/s41870-021-00817-5.
Zhang S, Zhang S, Zhang C, Wang X, Shi Y. Cucumber leaf disease identification with global pooling dilated convolutional neural network. Comput Electron Agric. 2019;162:422–430. https://doi.org/10.1016/j.compag.2019.03.012.
Sunil SH, Jayashri MR, Veena IP, Ayesha S, Pramodhini R. Plant leaf disease detection using computer vision and machine learning algorithms. Glob Trans Proc. 2022;3(1):305–10.
Barbedo JGA. Impact of dataset size and variety on the effectiveness of deep learning and transfer learning for plant disease classification. Comput Electron Agric. 2018;153:46–53. https://doi.org/10.1016/j.compag.2018.08.013.
Kavala SAPN, Pothuraju R. Detection of grape leaf disease using transfer learning methods: VGG16 & VGG19. In: 6th International Conference on Computing Methodologies and Communication (ICCMC), pp. 1205–1208. 2022. doi: https://doi.org/10.1109/ICCMC53470.2022.9753773.
Latif G, Abdelhamid SE, Mallouhy RE, Alghazo J, Kazimi ZA. Deep learning utilization in agriculture: detection of rice plant diseases using an improved CNN model. Plants. 2022;11:2230. https://doi.org/10.3390/plants11172230.
Mathulaprangsan S, Lanthong K, Jetpipattanapong D, Sateanpattanakul S, Patarapuwadol S. Rice diseases recognition using effective deep learning models, pp. 386–389. 2020. Doi: https://doi.org/10.1109/ECTIDAMTNCON48261.2020.9090709.
Sunil CK, Jaidhar CD, Patil N. Cardamom Plant Disease Detection Approach Using EfficientNetV2. In: IEEE Access, vol. 10, pp. 789–804. 2021. doi: https://doi.org/10.1109/ACCESS.2021.3138920.
Gupta S, Vishnoi J, Rao AS. Disease detection in maize plant using deep convolutional neural network. In: 7th International Conference on Communication and Electronics Systems (ICCES), pp. 1330–1335. 2022. doi: https://doi.org/10.1109/ICCES54183.2022.9835737.
Karlekar A, Seal A. SoyNet: Soybean leaf diseases classification. Comput Electron Agric. 2020;172:105342. https://doi.org/10.1016/j.compag.2020.105342.
Wang Y, Ke S, Wang S, Zheng Z. A grapevine virus disease detection method based on convolution neural network. In: 3rd International Conference on Computer Vision, Image and Deep Learning and International Conference on Computer Engineering and Applications (CVIDL & ICCEA), pp. 36–40. 2022 doi: https://doi.org/10.1109/CVIDLICCEA56201.2022.9825086
Turkoglu M, Hanbay D, Sengur A. (2022) Multi-model LSTM-based convolutional neural networks for detection of apple diseases and pests. J Ambient Intell Human Comput. 2022;13:3335–45. https://doi.org/10.1007/s12652-019-01591-w.
Akshay KP, Anitha J et al (2021) Plant disease classification using deep learning 2021 3rd International Conference on Signal Processing and Communication (ICPSC), pp. 407–411. 2021. doi: https://doi.org/10.1109/ICSPC51351.2021.9451696.
Hughes DP, Salathé M (2015) An open access repository of images on plant health to enable the development of mobile disease diagnostics. arXiv:1511.08060
Saleem MH, Khanchi S, Potgieter J, Arif KM. Image-based plant disease identification by deep learning meta-architectures. MDPI Plants. 2020;9:1451. https://doi.org/10.3390/plants9111451.
Chen J, Chen J, Zhang D, Sun Y, Nanehkaran YA. Using deep transfer learning for image-based plant disease identification. Comput Electron Agric. 2020;173:105393. https://doi.org/10.1016/j.compag.2020.105393.
Punam B, Pushkar G. Plant disease detection using hybrid model based on convolutional autoencoder and convolutional neural network. Artif Intell Agric. 2021;5:90–101. https://doi.org/10.1016/j.aiia.2021.05.002.
Pardede HF, Suryawati E, Sustika R, Zilvan V. Unsupervised convolutional autoencoder-based feature learning for automatic detection of plant diseases. In: International Conference on Computer, Control, Informatics and its Applications (IC3INA), pp. 158–162. 2018. doi: https://doi.org/10.1109/IC3INA.2018.8629518.
Benfenati A, Causin P, Oberti R, et al. Unsupervised deep learning techniques for automatic detection of plant diseases: reducing the need of manual labeling of plant images. J Math Ind. 2023;13:5. https://doi.org/10.1186/s13362-023-00133-6.
Abebech JB, Ayodeji OS, Minale A, Melaku BH. Development of a chickpea disease detection and classification model using deep learning. Inform Med Unlocked. 2022;31:100970. https://doi.org/10.1016/j.imu.2022.100970.
Turkoglu M, Yanikoğlu B. PlantDiseaseNet:convolutionalneural network ensemble for plant disease and pest detection. SIViP. 2022;16:301–9. https://doi.org/10.1007/s11760-021-01909-2.
Ruoling D, et al. Automatic diagnosis of rice diseases using deep learning. Front Plant Sci. 2021. https://doi.org/10.3389/fpls.2021.701038.
Liu B, Tan C, Li S, He J, Wang H. A data augmentation method based on generative adversarial networks for grape leaf disease identification. IEEE Access. 2020;8:102188–98. https://doi.org/10.1109/ACCESS.2020.2998839.
Yousef MAA, Orlando JMC, Liz MRB, Chamandeep K, Mohammed SAA, Bala BK. Leaf disease identification and classification using optimized deep learning. Meas Sens. 2023;25:100643. https://doi.org/10.1016/j.measen.2022.100643.
Li X, Li S. Transformer help CNN see better: a lightweight hybrid apple disease identification model based on transformers. Agriculture. 2022;12:884. https://doi.org/10.3390/agriculture12060884.
Sethy PK. Rice leaf disease image samples. Mendeley Data, V1. 2020. doi: https://doi.org/10.17632/fwcj7stb8r.1.
Shayanriyaz. 2020. https://www.kaggle.com/code/shayanriyaz/rice-crop-disease-detection-using-tensorflow/data. Accessed 1 Apr 2022
Fuentes A. Deep learning-based techniques for plant diseases recognition in real-field scenarios. In: Jacques B-T, Patrice D, Wilfried P, Dan P, Paul S, editors. Advanced concepts for intelligent vision systems. Cham: Springer; 2020. p. 3–14.
Caruana R, Niculescu-Mizil A, Crew G, Ksikes A (2004) Ensemble selection from libraries of models. In: Proceedings, Twenty-First International Conference on Machine Learning, ICML. New York: ACM Press; 2004. p. 137–144. doi: https://doi.org/10.1145/1015330.1015432.
Chen SL, Shen SQ, Li DS. Ensemble learning method for imbalanced data based on sample weight updating. Comput Sci. 2018;45(07):31–7.
El Asnaoui K. Design ensemble deep learning model for pneumonia disease classification. Int J Multimed Info Retr. 2021;10:55–68. https://doi.org/10.1007/s13735-021-00204-7.
Baccouche A, Begonya G-Z, Cristian CO, Adel E. Ensemble deep learning models for heart disease classification: a case study from Mexico. Information. 2020;11(4):207. https://doi.org/10.3390/info11040207.
Tao Z, Huiling L, Zaoli Y, Shi Q, Bingqiang H, Yali D. The ensemble deep learning model for novel COVID-19 on CT images. Appl Soft Comput. 2021;98:106885. https://doi.org/10.1016/j.asoc.2020.106885.
Patidar S, Pandey A, Shirish B, Sriram A. Rice plant disease detection and classification using deep residual learning. Proceedings. 2020. https://doi.org/10.1007/978-981-15-6315-7_23.
Prottasha SI, Reza SMS. A classification model based on depthwise separable convolutional neural network to identify rice plant diseases. Int J Electr Comput Eng. 2022;12(4):3642.
Lamba S, Baliyan A, Kukreja V. A novel GCL hybrid classification model for paddy diseases. Int J Inf Technol. 2022;15:1–10.
Orchi H, Sadik M, Khaldoun M. A novel hybrid deep learning model for crop disease detection using BEGAN. In: Sabir E, Elbiaze H, Falcone F, Ajib W, Sadik M, editors. Ubiquitous networking. UNet 2022. Lecture notes in computer science, vol. 13853. Cham: Springer; 2023.
Pandian JA, Kanchanadevi K, Kumar VD, Jasinska E, Gono R, Leonowicz Z, Jasinski MA. Five convolutional layer deep convolutional neural network for plant leaf disease detection. Electronics. 2022;11:1266.
Pradhan P, Kumar B, Mohan S. Comparison of various deep convolutional neural network models to discriminate apple leaf diseases using transfer learning. J Plant Dis Prot. 2022;129:1461–73. https://doi.org/10.1007/s41348-022-00660-1.
Sharada PM, David H, Marcel S. Using deep learning for image-based plant disease detection. Front Plant Sci. 2016. https://doi.org/10.48550/arXiv.1604.03169.
Chollet F. Xception deep learning with depthwise separable convolutions. In: IEEE conference on computer vision and pattern recognition (CVPR), Honolulu, HI, USA. 2017; p. 1800–7, https://doi.org/10.1109/CVPR.2017.195.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
There is no conflict of interests regarding the publication of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bathe, K., Patil, N., Patil, S. et al. ConvDepthTransEnsembleNet: An Improved Deep Learning Approach for Rice Crop Leaf Disease Classification. SN COMPUT. SCI. 5, 436 (2024). https://doi.org/10.1007/s42979-024-02783-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42979-024-02783-8