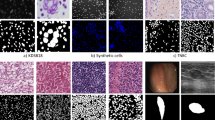
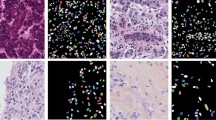
Abstract
The segmentation and classification of nuclei in haematoxylin and eosin-stained images is critical for diagnosing cancer and other disorders. Developing automated methods is necessary for the quantitative analysis of whole-slide images and further downstream analysis. However, many challenges are to be solved, such as varying morphology and observer differences. To address these concerns, we present ANet, an encoder–decoder structure based on attention mechanisms for nuclear segmentation and classification that makes use of information in high-dimensional features improved by attention. These blocks generate meaningful feature activation and eliminate irrelevant information to produce finer maps. It segments the touching, clustered, and overlapping nuclei and classifies them using upsampling branches. Our method includes components such as PreAct-ResNet50, residual attention, convolutional block attention module, and dense attention unit. We demonstrate how our approach achieves cutting-edge performance on several multi-tissue histopathology datasets such as Kumar, CoNSeP, and CPM17. We also demonstrate our model’s generalization capabilities on other combinations of datasets, including CPM15 and TNBC. ANet demonstrates a notable improvement of 1.14%, 2.70%, 1.41%, and 1.29% in Dice, AJI, SQ, and PQ scores, respectively, for the CPM17 dataset. In addition, it achieves a 1.18% improvement in AJI score for the Kumar dataset. Despite the inherent challenges in nuclei classification within the CoNSeP dataset, ANet yields outstanding results, showcasing a substantial improvement of 9.74%, 3.97%, and 0.80% in F1 scores for the inflammatory, spindle, and miscellaneous classes. Furthermore, ANet exhibits strong generalization across the CPM dataset, TNBC, and Combined CoNSeP, with improvements observed in the majority of metrics. The given improvement is justifiable, as are the interpretable visual results. The proposed method is of great potential for analyzing histopathology images, demonstrated by an increment of performance in multiple metrics.











Similar content being viewed by others
Data availability
The data supporting the findings of this study are available upon request from the corresponding author.
References
Gurcan MN, Boucheron LE, Can A, Madabhushi A, Rajpoot NM, Yener B. Histopathological image analysis: a review. IEEE Rev Biomed Eng. 2009;2:147–71. https://doi.org/10.1109/RBME.2009.2034865.
Graham S, Vu QD, Raza SEA, Azam A, Tsang YW, Kwak JT, et al. Hover-Net: simultaneous segmentation and classification of nuclei in multi-tissue histology images. Med Image Anal. 2019;58:101563. https://doi.org/10.1016/j.media.2019.101563.
Chen Y, Jia Y, Zhang X, Bai J, Li X, Ma M, Sun Z, Pei Z. TSHVNet: simultaneous nuclear instance segmentation and classification in histopathological images based on multiattention mechanisms. Biomed Res Int. 2022;2022:7921922. https://doi.org/10.1155/2022/7921922.
Olaf R, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells WM, Frangi AF, editors. Medical image computing and computer-assisted intervention – MICCAI 2015. Cham: Springer International Publishing; 2015. p. 234–41.
Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). 2015. pp. 3431–40. https://doi.org/10.1109/CVPR.2015.7298965.
Jha D, Smedsrud PH, Riegler MA, Johansen D, Lange T De, Halvorsen P, et al. ResUNet++: an advanced architecture for medical image segmentation. 2019 IEEE International Symposium on Multimedia (ISM). 2019. pp. 225–2255. https://doi.org/10.1109/ISM46123.2019.00049.
Shuttleworth JK, Todman AG, Naguib RNG, Newman BM, Bennett MK. Multiresolution colour texture analysis for classifying colon cancer images. Proceedings of the Second Joint 24th Annual Conference and the Annual Fall Meeting of the Biomedical Engineering Society] [Engineering in Medicine and Biology, vol. 2. 2002. pp. 1118–9 vol.2. https://doi.org/10.1109/IEMBS.2002.1106305.
LaTorre A, Alonso-Nanclares L, Muelas S, Peña JM, DeFelipe J. Segmentation of neuronal nuclei based on clump splitting and a two-step binarization of images. Expert Syst Appl. 2013;40:6521–30. https://doi.org/10.1016/j.eswa.2013.06.010.
Cheng J, Rajapakse JC. Segmentation of clustered nuclei with shape markers and marking function. IEEE Trans Biomed Eng. 2009;56:741–8. https://doi.org/10.1109/TBME.2008.2008635.
Zhou Z, Rahman Siddiquee MM, Tajbakhsh N, Liang J. UNet++: A Nested U-Net Architecture for Medical Image Segmentation. In: Stoyanov D, Taylor Z, Carneiro G, Syeda-Mahmood T, Martel A, Maier-Hein L, Tavares JMRS, Bradley A, Papa JP, Belagiannis V, Nascimento JC, Lu Z, Conjeti S, Moradi M, Greenspan H, Madabhushi A editors. Deep learning in medical image analysis and multimodal learning for clinical decision support. DLMIA ML-CDS 2018 2018. Lecture notes in computer science, vol 11045. Cham: Springer; 2018. https://doi.org/10.1007/978-3-030-00889-5_1
Xiao X, Lian S, Luo Z, Li S. Weighted Res-UNet for High-Quality Retina Vessel Segmentation. 2018 9th International Conference on Information Technology in Medicine and Education (ITME). 2018. pp. 327–31. https://doi.org/10.1109/ITME.2018.00080.
Raza SEA, Cheung L, Shaban M, Graham S, Epstein D, Pelengaris S, et al. Micro-Net: a unified model for segmentation of various objects in microscopy images. Med Image Anal. 2019;52:160–73. https://doi.org/10.1016/j.media.2018.12.003.
He H, Zhang C, Chen J, Geng R, Chen L, Liang Y, et al. A hybrid-attention nested UNet for nuclear segmentation in histopathological images. Front Mol Biosci. 2021. https://doi.org/10.3389/fmolb.2021.614174.
Chen H, Qi X, Yu L, Heng P-A. DCAN: Deep Contour-Aware Networks for Accurate Gland Segmentation. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). 2016. pp. 2487–96. https://doi.org/10.1109/CVPR.2016.273.
Doan TNN, Song B, Vuong TTL, Kim K, Kwak JT. SONNET: a self-guided ordinal regression neural network for segmentation and classification of nuclei in large-scale multi-tissue histology images. IEEE J Biomed Health Inform. 2022;26:3218–28. https://doi.org/10.1109/JBHI.2022.3149936.
Alashban A, Alsadan A, Alhussainan NF, Ouni R. Single convolutional neural network with three layers model for crowd density estimation. IEEE Access. 2022;10:63823–33. https://doi.org/10.1109/ACCESS.2022.3180738.
He K, Gkioxari G, Dollár P, Girshick R. Mask R-CNN. 2017 IEEE International Conference on Computer Vision (ICCV). 2017. pp. 2980–8. https://doi.org/10.1109/ICCV.2017.322.
Sirinukunwattana K, Raza SEA, Tsang Y-W, Snead DRJ, Cree IA, Rajpoot NM. Locality sensitive deep learning for detection and classification of nuclei in routine colon cancer histology images. IEEE Trans Med Imaging. 2016;35:1196–206. https://doi.org/10.1109/TMI.2016.2525803.
Zhou Y, Onder OF, Dou Q, Tsougenis E, Chen H, Heng P-A. CIA-Net: robust nuclei instance segmentation with contour-aware information aggregation. In: Chung ACS, Gee JC, Yushkevich PA, Bao S, editors. Information processing in medical imaging. Cham: Springer International Publishing; 2019. p. 682–93.
Cheng J, Pan X, Hou F, Zhao B, Lin J, Liu Z, et al. A standardized pipeline for colon nuclei identification and counting challenge. 2022. https://doi.org/10.48550/ARXIV.2203.00171.
Nauyan RS, Fraz MM. Nuclei probability and centroid map network for nuclei instance segmentation in histology images. Neural Comput Appl. 2023;35:15447–60. https://doi.org/10.1007/s00521-023-08503-2.
Zhou Y, Wu Y, Wang Z, Wei B, Lai M, Shou J, et al. Cyclic learning: bridging image-level labels and nuclei instance segmentation. IEEE Trans Med Imaging. 2023;42:3104–16. https://doi.org/10.1109/TMI.2023.3275609.
Zhang Y, Qi Y, Qi X, Senhadji L, Wei Y, Chen F, et al. FedSODA: federated cross-assessment and dynamic aggregation for histopathology segmentation. 2023. https://doi.org/10.48550/arXiv.2312.12824.
Yuan Y, Failmezger H, Rueda OM, Ali HR, Gräf S, Chin S-F, et al. Quantitative image analysis of cellular heterogeneity in breast tumors complements genomic profiling. Sci Transl Med. 2012;4:157ra143-157ra143. https://doi.org/10.1126/scitranslmed.3004330.
Shabbeer SH, Ghosh S, Kishan Babu K, Ram Dubey S, Pulabaigari V, Mukherjee S. RCCNet: an efficient convolutional neural network for histological routine colon cancer nuclei classification. 2018 15th International Conference on Control, Automation, Robotics and Vision (ICARCV). 2018. pp. 1222–7. https://doi.org/10.1109/ICARCV.2018.8581147.
Zhao S, He Y, Qin J, Wang Z. A semi-supervised deep learning method for cervical cell classification. Anal Cell Pathol. 2022;2022:4376178. https://doi.org/10.1155/2022/4376178.
Pati P, Foncubierta-Rodríguez A, Goksel O, Gabrani M. Reducing annotation effort in digital pathology: a co-representation learning framework for classification tasks. Med Image Anal. 2021;67:101859. https://doi.org/10.1016/j.media.2020.101859.
Jia Y, Lu C, Li X, Ma M, Pei Z, Sun Z, et al. Nuclei instance segmentation and classification in histopathological images using a DT-Yolact. 2021 20th International Conference on Ubiquitous Computing and Communications (IUCC/CIT/DSCI/SmartCNS). 2021. pp. 414–20. https://doi.org/10.1109/IUCC-CIT-DSCI-SmartCNS55181.2021.00072.
Hyun-Jic O, Jeong W-K. DiffMix: diffusion model-based data synthesis for nuclei segmentation and classification in imbalanced pathology image datasets. In: Greenspan H, Madabhushi A, Mousavi P, Salcudean S, Duncan J, Syeda-Mahmood T, Taylor R, editors. Medical image computing and computer assisted intervention – MICCAI 2023. Cham: Springer Nature Switzerland; 2023. p. 337–45.
Vo VT-T, Kim S-H. Mulvernet: nucleus segmentation and classification of pathology images using the HoVer-Net and multiple filter units. Electronics (Basel). 2023. https://doi.org/10.3390/electronics12020355.
Kaiming H, Zhang X, Ren S, Sun J. Identity mappings in deep residual networks. In: Bastian L, Matas J, Sebe N, Welling M, editors. Computer vision – ECCV 2016. Cham: Springer International Publishing; 2016. p. 630–45.
Wang F, Jiang M, Qian C, Yang S, Li C, Zhang H, et al. Residual attention network for image classification. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). 2017. pp. 6450–8. https://doi.org/10.1109/CVPR.2017.683.
Sanghyun W, Park J, Lee J-Y, Kweon IS. CBAM convolutional block attention module. In: Vittorio F, Hebert M, Sminchisescu C, Weiss Y, editors. Computer vision—ECCV 2018. Cham: Springer International Publishing; 2018. p. 3–19.
Yi-de M, Qing L, Zhi-bai Q. Automated image segmentation using improved PCNN model based on cross-entropy. Proceedings of 2004 International Symposium on Intelligent Multimedia, Video and Speech Processing, 2004. 2004. pp. 743–6. https://doi.org/10.1109/ISIMP.2004.1434171.
Sudre CH, Li W, Vercauteren T, Ourselin S, Jorge CM, et al. Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations. In: Cardoso MJ, Arbel T, Carneiro G, Syeda-Mahmood T, Tavares JMRS, Moradi M, et al., editors. Deep learning in medical image analysis and multimodal learning for clinical decision support. Cham: Springer International Publishing; 2017. p. 240–8.
Vu QD, Graham S, To MNN, Shaban M, Qaiser T, Koohbanani NA, et al. Methods for segmentation and classification of digital microscopy tissue images. Front Bioeng Biotechnol. 2018. https://doi.org/10.48550/ARXIV.1810.13230.
Kumar N, Verma R, Sharma S, Bhargava S, Vahadane A, Sethi A. A dataset and a technique for generalized nuclear segmentation for computational pathology. IEEE Trans Med Imaging. 2017;36:1550–60. https://doi.org/10.1109/TMI.2017.2677499.
Naylor P, Laé M, Reyal F, Walter T. Segmentation of nuclei in histopathology images by deep regression of the distance map. IEEE Trans Med Imaging. 2019;38:448–59. https://doi.org/10.1109/TMI.2018.2865709.
Carass A, Roy S, Gherman A, Reinhold JC, Jesson A, Arbel T, et al. Evaluating white matter lesion segmentations with refined Sørensen-dice analysis. Sci Rep. 2020. https://doi.org/10.1038/s41598-020-64803-w.
Kirillov A, He K, Girshick R, Rother C, Dollár P. Panoptic segmentation. 2018. https://doi.org/10.48550/ARXIV.1801.00868.
Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, et al. Cell Profiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. https://doi.org/10.1186/gb-2006-7-10-r100.
Bankhead P, Loughrey MB, Fernández JA, Dombrowski Y, McArt DG, Dunne PD, et al. QuPath: open source software for digital pathology image analysis. Sci Rep. 2017;7:16878. https://doi.org/10.1038/s41598-017-17204-5.
Badrinarayanan V, Kendall A, Cipolla R. SegNet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans Pattern Anal Mach Intell. 2017;39:2481–95. https://doi.org/10.1109/TPAMI.2016.2644615.
Acknowledgements
We would like to express our gratitude to the Department of Information Technology at the National Institute of Technology Karnataka, Surathkal, for providing us with the tools to conduct this research and experiment.
Funding
No funding was received for this work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The corresponding author declares that there is no conflict of interest on behalf of all the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the topical collection “AI Based Internet of Healthcare: Analysis and Future Perspectives” guest edited by Diganta Sengupta, Debashis De and Prasenjit Bhadra.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kadaskar, M., Patil, N. ANet: Nuclei Instance Segmentation and Classification with Attention-Based Network. SN COMPUT. SCI. 5, 348 (2024). https://doi.org/10.1007/s42979-024-02661-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42979-024-02661-3