Abstract
COVID-19 is creating havoc on the lives of human beings all around the world. It continues to affect the normal lives of people. As number of cases are high, a cost effective and fast system is required to detect COVID-19 at appropriate time to provide the necessary healthcare. Chest X-rays have emerged as an easiest way to detect COVID-19 in no time as RT-PCR takes time to detect the infection. In this paper we propose a concatenation-based CNN model that will detect COVID-19 from chest X-rays. We have developed a multiclass classification problem which can detect and classify a chest X-ray image as either COVID + ve, or viral pneumonia, or normal. We have used chest X-rays collected from different open sources. To maintain class balancing, we took 500 images of COVID, 500 normal images, and 500 pneumonia images. We divided our dataset in training, validation, and test set in 70:10:20 ratio respectively. We used four CNNs as feature extractors from the images and concatenated their feature maps to get better efficiency of the network. After training our model for 5 folds, we have obtained around 96.31% accuracy, 95.8% precision, 92.99% recall, and 98.02% AUC. We have compared our work with state-of-the-art pretrained transfer learning algorithms and other state-of-the-art CNN models referred in different research papers. The proposed model (Concat_CNN) exhibits better accuracy than the state-of-the-art models. We hope our proposed model will help to classify chest X-rays effectively and help medical professionals with their treatment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Coronaviruses are a large group virus which cause respiratory distress. COVID-19 or Coronavirus 2 is a new strain of this virus group that was previously not found in human body. This new strain of virus is named as ‘COVID-19’ virus as the first case of this virus was found in 2019. According to WHO’s confirmed report, first case of novel coronavirus (n-COV) was detected in Wuhan City, the largest metropolitan area of Hubei province in China in December 2019 [1]. COVID-19 virus is a single strand RNA that causes moderate to severe respiratory infection. In case of mild to moderate infection, infected person will recover within 14 days. As SARS-COV-2 is contagious and can spread rapidly among proximity of infected person, around 195 million people have been infected and around 4.1 million people died due to COVID-19 as of July 2021 [2]. Since the inception of the disease, be it developed countries like USA, China, Italy, England etc. or developing countries or underdeveloped countries, all are suffering due to huge case load and going through shortage of healthcare equipment, facilities, and lack of proper diagnosis approach with effective efficiency. SARS-COV-2 spreads rapidly through droplet from mouth, nose from an infected person to an uninfected person. Four variants of this virus—alpha, beta, gamma, delta are marked as variants of concern, whereas Eta, Iota, Kappa, Lambda falls under variant of interest [3].
Clinical Detection of COVID-19 largely depends on a genetic test known as Reverse Transcription Polymerase Chain Reaction (RT-PCR). Nasopharyngeal swabs are taken from infected patient for diagnosis purpose and RNA of the virus is extracted [4]. It takes around 2 h for detection with long turnaround time. So, it is costly, time consuming and complicated. If the swabs are not collected properly its accuracy rate becomes exceptionally low and might show negative result though the person is positive [5]. Rapid antigen testing is an easy-to-use process with less infrastructure requirement and equipment. It checks the presence of one or more antibodies in the infected person’s body as a response against the virus. Though it is cost friendly, easy to perform, quick response, and able to detect active SARS-COV-2 virus, its sensitivity and specificity is much lower than regular RT-PCR test [6]. The best solution is use of radiographic images like chest X-rays and chest CT-Scans. The advantage of using radiographic images is its wide availability all over the world. It is cost effective, and diagnosis is done at the time of testing. The main concern while using chest X-ray images is availability of trained doctors or healthcare professional. Research on infected chest X-ray images has found that some unknown features like vague darkening spots or ground glass opacity helps in COVID-19 detection [7]. However, analyzing every chest X-ray individually at the time of pandemic will delay the detection process thereby delaying the treatment procedure. Moreover, in rural places, availability of efficient radiologist is a problem for timely intervention and detection. So, an automatic COVID detection system or rather a computer aided COVID detection system is an important requirement at present scenario.
While the pandemic is still creating havoc on mankind, different researchers have come up with different ideas and proposed different systems for early detection and prevention of diseases. Among them applications of deep learning algorithms have shown promising results. In healthcare sector, deep learning [8] has shown tremendous development in automatic disease prediction such as tuberculosis detection [9, 10], cancer detection [11, 12], tumor detection [13, 14], bone fracture detection [15, 16], genome sequence analysis [17,18,19] etc. In their study, Abbas et al. [20], proposed a model named DeTraC that classifies chest X-ray images. The model is based on deep CNN models. Class decomposition layer was used to simplify the dataset structure. ResNet pretrained model was used to pretrain the model and then class-composition layer was used for parameter fine tuning. The dataset size was 185 images where 105 images are virus infected and 80 are normal chest X-ray images. The authors have achieved 95.12% accuracy, 97.91% sensitivity and 91.87% specificity. Das et al. [21], had proposed an ensemble of multiple state of the art CNN models like InceptionV3, ResNet50V2, and DenseNet201. All these models are trained individually and then training results are combined using weighted average ensemble technique. The dataset containing 538 COVID + images and 468 COVID − images is divided into train, test and validation set. The accuracy of the proposed model is 91.62%. Wang et al. [22] proposed a custom deep learning architecture named COVIDNet that classifies images into three categories: COVID infected patients, healthy patients, and pneumonia patients. Their model is designed using a lightweight projection-expansion-projection-extension (PEPX) pattern which obtained a classification accuracy of 94%. Apostolopoulos et al. [23] proposed COVID-19 identification system using five pretrained deep learning models. Transfer learning models VGG19, Inception, Xception, MobileNet, and Inception_ResNet_V2 are used for classification. They achieved accuracy around 96.78%. Their binary classification results are better than multiclass classification. A hybrid model combining Xception and ResNet50_V2 model was proposed by Rahimzadeh et al. [24]. They used a dataset of 6054 X-ray images and achieved an accuracy of 91.4%. Fu et al. [25] explored two characteristics of SARS-COV-2-dynamic viral shedding and serological characteristics. They identified the risk factors associated with poor recovery and lung lesion resolutions. They studied the relationship between pro-inflammatory response, viral shedding, and lung lesion evolution. According to their study, 58% of the infected patients will have poor recovery and prolonged interval of viral shedding. In another study, Cheng et al. [26], compared the performances of different ensemble CNN models like majority voting, unweighted average, and Bayes Optimal Classifier. Afsar et al. [27], have proposed a model named COVID-CAAPS which is based on capsule network to detect COVID infection. They combined different convolution and capsule layers to address the problem of class-imbalance. With smaller number of trainable parameters, they obtained an accuracy of 95.7%, sensitivity of 90% and specificity of 95.8%. A model named COVID-RENet [28] was proposed by Cohen et al. which extracts edge and region-based features using CNN. Later SVM was used to perform classification and improve accuracy. They have implemented a fivefold cross validation for on the COVID-19 dataset.
Different researchers have proposed different models for COVID detection using deep learning approaches. Most of the works are based on CNN as it shows higher accuracy for classification due to its hierarchical structure.
We have implemented a concatenation based convolutional neural network model for COVID-19 detection from chest X-rays and compared our results with state-of-the-art benchmarked transfer learning algorithms and other state-of-the-art works done by different researchers. This paper is organized as follows-
“Data Description” of the paper discusses the details of the data used for implementation. It focuses about the type of data available, data preprocessing steps, and the tools and environment we used for our model implementation. “Proposed Model” presents the detailed architecture of our proposed model. It also provides the training details and parameter fine tuning details. “Implementation Results” shows the implementation results using different performance metrics and comparisons are shown with state-of-the art transfer learning models and other state-of-the-art other research works. Finally, “Conclusion” concludes the implementation and discussion on the outcome of the proposed model.
Data Description
In this section details of the data source, dataset description, image preprocessing techniques are discussed.
Data Collection
For performing our proposed research work, we collected chest X-ray images from different online open sources. These dataset sources contain chest X-ray images having different infection manifestations including COVID-19 viral infection. These images are collected from all over the world and contain different chest X-ray images like viral pneumonia, bacterial pneumonia, normal chest X-ray images. Chest X-ray images of Viral infections like COVID-19, SARS, MERS-CoV, varicella, influenza, herpes and bacterial infections like streptococcus, staphylococcus, Klebsiella, Escherichia coli, Mycoplasma are included in these datasets with detailed medical information. Different views of these images, like: anterior–posterior view (AP), lateral view (L), and posterior anterior (PA) view are available. As our focus is to view the condition of the lungs for any infection, posterior-anterior (PA) views suit best. For this purpose, we have considered images having PA views only and discarded other views. Even though there are many viral and bacterial categories of images, for our experiment purpose we have considered three classes—COVID-19 positive, Normal chest X-ray, and viral pneumonia. Normal chest X-ray (CXR) images are considered as class 0, COVID-19 positive as class 1, and viral pneumonia as class 2. To maintain class balancing, we have created our dataset with equal number of images in each class, i.e., 500 COVID Chest X-ray images, 500 normal chest X-ray images, and 500 chest X-ray images of viral pneumonia. We have obtained our images from the following open sources:
-
i.
150 images from GitHub which has images from all three classes i.e., 50 images from each class (https://github.com/ieee8023/covid-chestxray-dataset).
-
ii.
55 COVID images from GitHub (https://github.com/Figure-1-COVID-chestxray-dataset).
-
iii.
250 COVID images, 300 normal images and 270 Pneumonia images (https://www.kaggle.com/tawsifurrahman/covid19-radiography-database).
-
iv.
95 COVID images and 75 normal images(https://www.kaggle.com/nabeelsajid917/covid-19-x-ray-10000-images).
-
v.
75 normal images and 230 pneumonia images (https://www.kaggle.com/paulimothymooney/chest-xray-pneumonia).
Data Preprocessing
As the images are collected from different sources, images are of different shapes and their brightness and contrast are different. So, the images are first resized to 224 × 224 shape. To improve the contrast of all images, we implemented an image processing technique Contrast Limited Adaptive Histogram Equalization technique (CLAHE) [39]. Here the entire image is divided into fixed number of tiles, contrast of each tile is enhanced, and then bilinear interpolation is used to remove the artificial boundaries within the images. CLAHE is used to enhance the lungs region of each image for better understanding. The images are next normalized by a factor of 1/255. The images are then shuffled for better distribution and the whole dataset is divided into train, validation, and test set in a ratio of 70:10:20 respectively (Table 1). In Table 2, a clear insight of image data distribution in provided.
Tools Used for Implementation
We have used Google Colab GPU (Tesla K80), Python 3.7. We used TensorFlow 2.7.0 and Keras 2.7.0 libraries for our model implementation on Google COLAB platform (Table 2).
Proposed Model
We have implemented a concatenation approach of convolutional neural network for COVID-19 detection using chest X-rays. Concatenation of CNNs’ helps to combine the features extracted by CNNs’ to enhance the classification process more accurate. Four different CNNs with different convolutional layer architecture are implemented as feature extractors from the training dataset. This section discusses the proposed concatenated CNN architecture, necessary optimization process, and parameter fine tuning stages.
CNNs as Feature Extractor
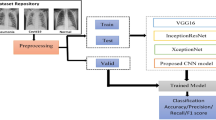
In this work four CNN architectures are used as feature extractors. These CNNs have been used to learn good representations of the images like edges, shapes, rotation, spatial representation. These models extract relevant features from the chest X-ray images to obtain a proper knowledge about normal, COVID infected and pneumonia characteristics. We have used Scikit-Learn, Keras, TensorFlow, OpenCV python libraries for implementing different functionalities on the model. Below image shows the architecture illustration of our COVID detection model.
In our proposed model in Fig. 1, we have illustrated four different CNN architectures. The preprocessed training images of size 224 × 224 pixel are provided as input to all four CNNs. Feature maps are generated from the last feature extraction layer of all extractors. As all CNNs extract same size of feature maps, we use a concatenation function to generate an enhanced quality of semantic features. A concatenated convolutional neural network (Concat_CNN) is proposed by using all extracted feature maps concatenation and connecting this concatenation layer to a fully connected layer with 32 neurons in association with ReLU Activation function which is further connected to a classifier layer. Our proposed model is implemented as a multi-class classification problem as we have categorized our data in three classes as COVID-19, normal, and pneumonia. Each feature extractor CNN is composed of two convolutional layers with varying filter size of 3 × 3. Each convolution layer is followed by a batch normalization layer, maxpooling layer with pool size (2 × 2) followed by a dropout rate at 0.5. ReLU activation function is used in each convolution layer. Rectified activation function is one of the most popular activation functions which directly maps the inputs to the outputs if it is positive, else it maps zero. It performs better than vanishing gradient and works faster. The equation of the Relu function can be defined as:
Keras concatenation function concatenate is used to merge all feature extractors output. An interpretation layer or a fully connected layer with 32 neurons is further added followed by the final output layer. Activation function Softmax is used in the output layer as it acts best in multiclass classification problem. The equation of Softmax function can be defined as:
In each convolutional layer, a kernel initializer ‘he_uniform’ is used.
Kernel initializers are used to stabilize the weight initialization and distribution for neural network in each stage of iteration. Below tables provide an insight of the CNNs used as the feature extractors (Tables 3, 4, 5, 6).
Training and Parameter Fine Tuning
The proposed model is compiled using Adam optimizer provided by Keras library with the parameters as learning rateα α = 0.0001. Categorical Crossentropy Loss function is used as the model is a multiclass classification problem. The model has been trained for 50 epochs with Early stopping callbacks with patience for 10 epochs. To train our model effectively, we have considered fivefold cross validation in addition to splitting of images into different sets. In each fold, a different set of images is used as training and testing set while keeping the validation image set fixed.
Implementation Results
Performance Metrics
We evaluate our proposed approach by considering the following four performance evaluation metrics—classification accuracy, precision, recall, AUC [35].
Where TP stands for True Positive, TN stands for True Negative, FP stands for False Positive, FN stands False Negative. COVID + cases that are correctly classified are called True positive cases in the confusion matrix. If COVID + cases are falsely classified as COVID − then these are called False Positive. Same wise, if COVID − cases are classified correctly then these are called True Negative and COVID − cases are falsely classified as COVID + are known as False Negative.
Performance Evaluation and Compare Benchmark
In our work, we have proposed a concatenation-based CNN (Concat_CNN) model for COVID-19 disease prediction. However, for comparison purpose we have compared our proposed model results with other contemporary works done for COVID-19 disease prediction using chest X-rays. We also compared our work with individual transfer learning algorithms performance. This provides a better understanding of our proposed model’s performance with other contemporary works and benchmark transfer learning algorithms. As our model is trained with fivefold cross validation and for 50 epochs with learning rate at 0.0001 and Adam optimizer, same training approach is adopted for all transfer learning algorithm implementation. We have considered VGG16 [31], InceptionV3 [34], ResNet50 [32], and DenseNet201 [33] transfer learning algorithms. All model evaluation results are saved as.h5 files.
Table 7 demonstrates an overview of accuracy, precision, recall and AUC of the benchmark transfer learning algorithms and our proposed model. It is clearly understood from the table that our proposed model exhibits better results in most of the folds in terms of accuracy, precision, recall and AUC. Average accuracy obtained by our model is 96.31%, average precision is 95.8%, average Recall is 92.99% and average AUC is 98.02%. It helps to prove the efficiency and robustness of the proposed approach.
Figure 2 shows the confusion matrix of our proposed model obtained in all five folds. Each confusion matrix provides an overview of consistent performance of our model in all folds.
If we analyze the confusion matrix of all folds, our concatenated CNN model detects most of the cases correctly. From this confusion matrix, it is evident that the model clearly identified true positive and true negative values efficiently.
In Fig. 3, accuracy, loss, precision, recall and AUC graphs are shown for all five folds. In each fold all the metrics exhibit better results than the previous folds. The proposed model also gives consistent high precision which again proves the sturdiness of the model.
Also, to demonstrate our proposed concatenated CNN model’s efficacy towards detection of COVID cases based on chest X-rays, a comparison is made in Table 8 with some recent studies and their implementation details. Among the different research works, some have shown promising results whereas some have faced difficulty with a small dataset [37, 38]. Some have even proposed new methods which shows promising results [37, 38]. From the Table 8 it can be observed that our proposed Concat_CNN model has the capability to yield better results than state-of-the-art deep learning models [24, 29, 30, 36] and implementations.
Conclusion
In our paper, we have implemented a concatenation-based CNN (Concat_CNN) model to classify chest X-ray images into three classes: normal, COVID, and pneumonia. We used four CNN networks as feature extractors and concatenated the feature map to get more accurate classification. We obtained our dataset from different open sources. We tried to maintain class balancing by choosing equal no. of images for all classes in training phase. Our proposed model has an average classification accuracy of 96.31% between five folds. We hope our model will be helpful for medical diagnosis as number of COVID cases are still significantly increasing. So, a computer based COVID detection system will accelerate the detection and classification and will further help to solve the case load problem of medical professionals.
References
Rehman MFU, Fariha C, Anwar A, Shahzad N, Ahmad M, Mukhtar S, Farhan Ul Haque M. Novel coronavirus disease (COVID-19) pandemic: a recent mini review. Comput Struct Biotechnol J. 2021;19:612–23. https://doi.org/10.1016/j.csbj.2020.12.033 (Epub 2020 Dec 31).
https://www.who.int/publications/m/item/weekly-operational-update-on-covid-19---26-july-2021
https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/
Bustin SA. Absolute quantification of mRNA using realtime reverse transcription polymerase chain reaction assays. J Mol Endocrinol. 2000;25(2):169–93.
Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, Wang B, Xiang H, Cheng Z, Xiong Y, Zhao Y, Li Y, Wang X, Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 Novel Coronavirus-infected pneumonia in Wuhan, China. J Am Med Assoc. 2020. https://doi.org/10.1001/jama.2020.1585.
Xia W, Shao J, Guo Y, Peng X, Li Z, Hu D. Clinical and CT features in pediatric patients with COVID-19 infection: different points from adults. Pediatr Pulmonol. 2020;55:1169–74. https://doi.org/10.1002/ppul.24718.
Hesamian MH, Jia W, He X, et al. Deeplearning techniques for medical image segmentation: achievements and challenges. J Digit Imaging. 2019;32:582–96. https://doi.org/10.1007/s10278-019-00227-x.
Oloko-Oba M, Viriri S. Diagnosing tuberculosis using deep convolutional neural network. In: El Moataz A, Mammass D, Mansouri A, Nouboud F, editors. Image and signal processing. ICISP 2020. Lecture notes in computer science, vol. 12119. Cham: Springer; 2020. https://doi.org/10.1007/978-3-030-51935-3_16.
Dinesh Jackson Samuel R, Rajesh Kanna B. Tuberculosis (TB) detection system using deep neural networks. Neural Comput Appl. 2019;31:1533–45. https://doi.org/10.1007/s00521-018-3564-4.
Dabral I, Singh M, Kumar K. Cancer detection using convolutional neural network. In: Tripathi M, Upadhyaya S, editors. Conference proceedings of ICDLAIR2019. ICDLAIR 2019. Lecture notes in networks and systems, vol. 175. Cham: Springer; 2021. https://doi.org/10.1007/978-3-030-67187-7_30.
Sharma A, Kochar B, Joshi N, Kumar V. Breast cancer detection using deep learning and machine learning: a comparative analysis. In: Gupta D, Khanna A, Bhattacharyya S, Hassanien AE, Anand S, Jaiswal A, editors. International conference on innovative computing and communications. Advances in intelligent systems and computing, vol. 1165. Singapore: Springer; 2021. https://doi.org/10.1007/978-981-15-5113-0_39.
Irmak E. Multi-Classification of Brain Tumor MRI images using deep convolutional neural network with fully optimized framework. Iran J Sci Technol Trans Electr Eng. 2021;45:1015–36. https://doi.org/10.1007/s40998-021-00426-9.
More RB, Bhisikar SA. Brain tumor detection using deep neural network. In: Pawar PM, Balasubramaniam R, Ronge BP, Salunkhe SB, Vibhute AS, Melinamath B, editors. Techno-societal 2020. Cham: Springer; 2021. https://doi.org/10.1007/978-3-030-69921-5_9.
Kitamura G, Chung C, Moore BE. Ankle fracture detection utilizing a convolutional neural network ensemble implemented with a small sample, de novo training, and multiview incorporation. J Digit Imaging. 2019;32:672–7. https://doi.org/10.1007/s10278-018-0167-7.
Yadav DP, Rathor S (2020) Bone fracture detection and classification using deep learning approach. In: International Conference on Power Electronics & IoT Applications in Renewable Energy and its Control (PARC). pp 282–285.https://doi.org/10.1109/PARC49193.2020.236611.
Liu J, Li J, Wang H, et al. Application of deep learning in genomics. Sci China Life Sci. 2020;63:1860–78. https://doi.org/10.1007/s11427-020-1804-5.
Danilevsky A, Shomron N. Deep learning applied on next generation sequencing data analysis. In: Shomron N, editor. Deep sequencing data analysis. Methods in molecular biology, vol. 2243. New York, NY: Humana; 2021. https://doi.org/10.1007/978-1-0716-1103-6_9.
Kotlarz K, Mielczarek M, Suchocki T, et al. The application of deep learning for the classification of correct and incorrect SNP genotypes from whole-genome DNA sequencing pipelines. J Appl Genetics. 2020;61:607–16. https://doi.org/10.1007/s13353-020-00586-0.
Abbas A, Abdelsamea MM, Gaber MM. Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. Appl Intell. 2021;51:854–64. https://doi.org/10.1007/s10489-020-01829-7.
Das AK, Ghosh S, Thunder S, et al. Automatic COVID-19 detection from X-ray images using ensemble learning with convolutional neural network. Pattern Anal Applic. 2021. https://doi.org/10.1007/s10044-021-00970-4.
Wang L, Lin ZQ, Wong A. COVID-Net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. Sci Rep. 2020;10:19549. https://doi.org/10.1038/s41598-020-76550-z.
Apostolopoulos ID, Mpesiana TA. Covid-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med. 2020;43:635–40. https://doi.org/10.1007/s13246-020-00865-4.
Rahimzadeh M, Abolfazl Attar A. A modified deep convolutional neural network for detecting COVID-19 and pneumonia from chest X-ray images based on the concatenation of Xception and ResNet50V2. Inf Med Unlocked. 2020;19:100360. https://doi.org/10.1016/j.imu.2020.100360.
Fu S, Fu X, Song Y, Li M, Pan PH, Tang T, Zhang C, Jiang T, Tan D, Fan X, Sha X. Virologic and clinical characteristics for prognosis of severe COVID-19: a retrospective observational study in Wuhan, China. medRxiv. 2020;395:507. https://doi.org/10.1101/2020.04.03.20051763.
Ju C, Bibaut A, van der Laan M. The relative performance of ensemble methods with deep convolutional neural networks for image classification. J Appl Stat. 2018;45(15):2800–18. https://doi.org/10.1080/02664763.2018.1441383.
Afshar P, Heidarian S, Naderkhani F, Oikonomou A, Plataniotis KN, Mohammadi A (2020) Covid-caps: a capsule network-based framework for identification of COVID-19 cases from X-ray images. arXiv preprint arXiv:2004.02696
Cohen JP, Morrison P, Dao L (2020) COVID-19 image data collection. arXiv preprint arXiv:2003.11597
Zebin T, Rezvy S. COVID-19 detection and disease progression visualization: Deep learning on chest X-rays for classification and coarse localization. Appl Intell. 2021;51:1010–21. https://doi.org/10.1007/s10489-020-01867-1.
Ibrahim AU, Ozsoz M, Serte S, et al. Pneumonia classification using deep learning from chest X-ray images during COVID-19. Cogn Comput. 2021. https://doi.org/10.1007/s12559-020-09787-5.
Simonyan K, Zisserman A (2014) Very Deep convolutional networks for large-scale image recognition. arXiv:1409.1556.
He K, Zhang X, Ren S, Sun J (2015) Deep residual learning for image recognition. 7.
Huang G, Liu Z, van der Maaten L, Weinberger KQ. Densely connected convolutional networks. In: 2017 IEEE conference on computer vision and pattern recognition (CVPR). IEEE; 2017, pp 2261–2269. https://doi.org/10.1109/CVPR.2017.243.
Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D et al (2015) Going deeper with convolutions. In: IEEE Conference on Computer Vision and Pattern Recognition (CVPR); Boston, MA, pp 19.
Sokolova M, Lapalme G. A systematic analysis of performance measures for classification tasks. Inf Process Manag. 2009;45(4):427–37.
Das AK, Ghosh S, Thunder S, et al. Automatic COVID-19 detection from X-ray images using ensemble learning with convolutional neural network. Pattern Anal Applic. 2021;24:1111–24. https://doi.org/10.1007/s10044-021-00970-4.
Wang L, Lin ZQ, Wong A. COVID-Net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. Sci Rep. 2020;10(1):19549. https://doi.org/10.1038/s41598-020-76550-z.
Khan AI, Shah J, Bhat M. CoroNet: a deep neural network for detection and diagnosis of COVID-19 from chest X-ray images. Comput Methods Programs Biomed. 2020;196:105581 (Accessed 29 Mar 2020).
Reza AM. Realization of the contrast limited adaptive histogram equalization (CLAHE) for real-time image enhancement. J VLSI Signal Process Systr Signal, Image Video Technol. 2004;38:35–44. https://doi.org/10.1023/B:VLSI.0000028532.53893.82.
Funding
West Bengal State government fellowship scheme, India (Reference no. P-1/RS/57/19).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the topical collection “Social Data Science: Research Challenges and Future Directions” guest edited by Sarbani Roy, Chandreyee Chowdhury and Samiran Chattopadhyay.
Rights and permissions
About this article
Cite this article
Saha, P., Neogy, S. Concat_CNN: A Model to Detect COVID-19 from Chest X-ray Images with Deep Learning. SN COMPUT. SCI. 3, 305 (2022). https://doi.org/10.1007/s42979-022-01182-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42979-022-01182-1