Abstract
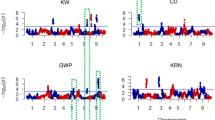
Maize, a one of the cereal crops, serves as the staple food for numerous people across the world. Maize breeding programmes primarily aim to enhance the yield potential of newly developed cultivars. This objective can be accomplished either by direct selection for increased grain yield or by indirect selection for traits closely associated with yield. The present study was carried out in two environments viz., Coimbatore (E1) and Vagarai (E2) to locate QTLs associated with yield-related traits in a recombinant inbred line (RIL) population that resulted from a cross between UMI79 and UMI 936(w). A total of seven yield-related traits (Ear weight, ear diameter, ear length, kernel row number, kernel number per row, 100-kernel weight, and grain yield) were evaluated across the two environments. The QTLs were mapped with the support of genotyping-by-sequencing based SNPs genetic linkage map which contains 1516 markers and spans 10 chromosomes and covers an overall distance of 6924.7 centimorgans. A total of 22 QTLs were discovered across these environments and best linear unbiased predictors, with four of these QTLs being common across them. The study pinpointed three stable QTLs (qEW-6, qEL-2, and qKRN-5), each contributing > 10% of the phenotypic variance in ear weight, ear length, and kernel row number, respectively. Within these QTL regions, 109 protein-coding genes were identified. Gene ontology analysis revealed these genes roles in development, reproduction, and growth. Several candidate genes, including MADS-box transcription factors, serine carboxypeptidase, and E3 ubiquitin-protein ligase, are well known for their role in yield-related traits of maize, and other cereal crops were among those identified. These findings provide valuable information for maize breeders aiming to enhance grain yield-related traits through marker-assisted breeding.





Similar content being viewed by others
References
Bar-Peled M, O’Neill MA (2011) Plant nucleotide sugar formation, interconversion, and salvage by sugar recycling. Annu Rev Plant Biol 62:127–155. https://doi.org/10.1146/annurev-arplant-042110-103918
Byrne S, Czaban A, Studer B, Panitz F, Bendixen C, Asp T (2013) Genome wide allele frequency fingerprints (GWAFFs) of populations via genotyping by sequencing. PLoS ONE 8(3):e57438. https://doi.org/10.1371/journal.pone.0057438
Cai M, Li S, Sun F, Sun Q, Zhao H, Ren X, Zhao Y, Tan BC, Zhang Z, Qiu F (2017) Emp10 encodes a mitochondrial PPR protein that affects the cis-splicing of nad2 intron 1 and seed development in maize. Plant J 91(1):132–144. https://doi.org/10.1111/tpj.13551
Chen Z, Wang B, Dong X, Liu H, Ren L, Chen J, Hauck A, Song W, Lai J (2014) An ultra-high density bin-map for rapid QTL mapping for tassel and ear architecture in a large F2 maize population. BMC Genomics 15:1–10. https://doi.org/10.1186/1471-2164-15-433
Dai D, Luan S, Chen X, Wang Q, Feng Y, Zhu C, Qi W, Song R (2018) Maize Dek37 encodes a P-type PPR protein that affects cis-splicing of mitochondrial nad2 intron 1 and seed development. Genetics 208(3):1069–1082. https://doi.org/10.1534/genetics.117.300602
Danilevskaya ON, Meng X, Selinger DA, Deschamps S, Hermon P, Vansant G, Gupta R, Ananiev EV, Muszynski MG (2008) Involvement of the MADS-box gene ZMM4 in floral induction and inflorescence development in maize. Plant Physiol 147(4):2054–2069. https://doi.org/10.1104/pp.107.115261
Davis G, McMullen M, Baysdorfer C, Musket T, Grant D, Staebell M, Xu G, Polacco M, Koster L, Melia-Hancock S (1999) A maize map standard with sequenced core markers, grass genome reference points and 932 expressed sequence tagged sites (ESTs) in a 1736-locus map. Genetics 152(3):1137–1172. https://doi.org/10.1093/genetics/152.3.1137
De Leon TB, Pruthi R, Jampala B, Borjas AH, Subudhi PK (2020) Genetic determinants for agronomic and yield-related traits localized on a GBS-SNP linkage map from a japonica x indica cross in rice. Plant Gene 24:100249. https://doi.org/10.1016/j.plgene.2020.100249
Devasree S, Ganesan K, Ravikesavan R, Senthil N, Paranidharan V (2020) Relationship between yield and its component traits for enhancing grain yield in single cross hybrids of maize (Zea mays L.). Electron J Plant Breed 11(3):796–802
Duvick DN (2005) The contribution of breeding to yield advances in maize (Zea mays L.). Adv Agron 86:83–145. https://doi.org/10.1016/S0065-2113(05)86002-X
Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6(5):e19379. https://doi.org/10.1371/journal.pone.0019379
Gallais A, Hirel B (2004) An approach to the genetics of nitrogen use efficiency in maize. J Exp Bot 55(396):295–306. https://doi.org/10.1093/jxb/erh006
Hasan N, Choudhary S, Naaz N, Sharma N, Laskar RA (2021) Recent advancements in molecular marker-assisted selection and applications in plant breeding programmes. J Genet Eng Biotechnol 19(1):1–26. https://doi.org/10.1186/s43141-021-00231-1
Helentjaris T, Slocum M, Wright S, Schaefer A, Nienhuis J (1986) Construction of genetic linkage maps in maize and tomato using restriction fragment length polymorphisms. Theor Appl Genet 72:761–769. https://doi.org/10.1007/BF00266542
Hussin SH, Wang H, Tang S, Zhi H, Tang C, Zhang W, Jia G, Diao X (2021) SiMADS34, an E-class MADS-box transcription factor, regulates inflorescence architecture and grain yield in Setaria italica. Plant Mol Biol 105:419–434. https://doi.org/10.1007/s11103-020-01097-6
IBPGR (1991) Descriptors for Maize, International Maize and Wheat Improvement Center Mexico City/International Board for Plant Genetic Resources, Rome
Jadhav KP, Senthil N, Tamilarasi PM, Ganesan KN, Paranidharan V, Raveendran M, Ramalingam J (2019) QTL mapping for sorghum downy mildew disease resistance in maize (Zea mays L.) in recombinant inbred line population of UMI79 X UMI936 (w). Curr Plant Biol 20:100124. https://doi.org/10.1016/j.cpb.2019.100124
Jadhav KP, Saykhedkar GR, Tamilarasi PM, Devasree S, Ranjani RV, Sarankumar C, Bharathi P, Karthikeyan A, Arulselvi S, Vijayagowri EJFIG (2022) GBS-based SNP map pinpoints the QTL associated with sorghum downy mildew resistance in maize (Zea mays L.). Front Genet 13:890133
Jiang P, Zhang H, Hao Z, Li B, Li M, Wang H, Ci X, Zhang S, Li X, Shi Z (2013) Analysis of meta-QTL and candidate genes related to yield components in maize. Acta Agron Sin 39(6):969–978. https://doi.org/10.3724/SP.J.1006.2013.00969
Kong W, An B, Zhang Y, Yang J, Li S, Sun T, Li Y (2019) Sugar transporter proteins (STPs) in Gramineae crops: comparative analysis, phylogeny, evolution, and expression profiling. Cells 8(6):560. https://doi.org/10.3390/cells8060560
Li Y, Fan C, Xing Y, Jiang Y, Luo L, Sun L, Shao D, Xu C, Li X, Xiao J (2011) Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat Genet 43(12):1266–1269. https://doi.org/10.1038/ng.977
Li Z, Tang L, Qiu J, Zhang W, Wang Y, Tong X, Wei X, Hou Y, Zhang J (2016) Serine carboxypeptidase 46 regulates grain filling and seed germination in rice (Oryza sativa L.). PLoS ONE 11(7):e0159737. https://doi.org/10.1371/journal.pone.0159737
Liu Y, Yi Q, Hou X, Hu Y, Li Y, Yu G, Liu H, Zhang J, Huang Y (2020) Identification of quantitative trait loci for kernel-related traits and the heterosis for these traits in maize (Zea mays L.). Mol Genet Genom 295:121–133. https://doi.org/10.1007/s00438-019-01608-1
Lu Y, Yan J, Guimaraes CT, Taba S, Hao Z, Gao S, Chen S, Li J, Zhang S, Vivek BS (2009) Molecular characterization of global maize breeding germplasm based on genome-wide single nucleotide polymorphisms. Theor Appl Genet 120:93–115. https://doi.org/10.1007/s00122-009-1162-7
Mena M, Mandel MA, Lerner DR, Yanofsky MF, Schmidt RJ (1995) A characterization of the MADS-box gene family in maize. Plant J 8(6):845–854. https://doi.org/10.1046/j.1365-313X.1995.8060845.x
Meng L, Li H, Zhang L, Wang J (2015) QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J 3(3):269–283. https://doi.org/10.1016/j.cj.2015.01.001
Nzuve F, Githiri S, Mukunya D, Gethi J (2014) Genetic variability and correlation studies of grain yield and related agronomic traits in maize. J Agric Sci 6(9):166
Panwar L, Mahawar R, Narolia R (2013) Genetic variability and interrelationships among grain yield and yield components in maize. Ann Plant Soil Res 15(1):15–18
Peterson BG, Carl P, Boudt K, Bennett R, Ulrich J, Zivot E, Cornilly D, Hung E, Lestel M, Balkissoon K (2018) Package ‘performanceanalytics.’ R Team Cooper 3:13–14
Popat R, Patel R, Parmar D (2020) Variability: genetic variability analysis for plant breeding research. R package version 01 0
Ren X, Pan Z, Zhao H, Zhao J, Cai M, Li J, Zhang Z, Qiu F (2017) EMPTY PERICARP11 serves as a factor for splicing of mitochondrial nad1 intron and is required to ensure proper seed development in maize. J Exp Bot 68(16):4571–4581. https://doi.org/10.1093/jxb/erx212
Saedler H, Becker A, Winter K-U, Kirchner C, Theißen G (2001) MADS-box genes are involved in floral development and evolution. Acta Biochim Pol 48(2):351–358
Senior M, Chin E, Lee M, Smith J, Stuber C (1996) Simple sequence repeat markers developed from maize sequences found in the GENBANK database: map construction. Crop Sci 36(6):1676–1683. https://doi.org/10.2135/cropsci1996.0011183X003600060043x
Shulga O, Shchennikova A, Angenent G, Skryabin K (2008) MADS-box genes controlling inflorescence morphogenesis in sunflower. Russ J Dev Biol 39:2–5. https://doi.org/10.1134/S1062360408010025
Sonah H, Bastien M, Iquira E, Tardivel A, Légaré G, Boyle B, Normandeau É, Laroche J, Larose S, Jean M (2013) An improved genotyping by sequencing (GBS) approach offering increased versatility and efficiency of SNP discovery and genotyping. PLoS ONE 8(1):e54603. https://doi.org/10.1371/journal.pone.0054603
Spindel J, Wright M, Chen C, Cobb J, Gage J, Harrington S, Lorieux M, Ahmadi N, McCouch S (2013) Bridging the genotyping gap: using genotyping by sequencing (GBS) to add high-density SNP markers and new value to traditional bi-parental mapping and breeding populations. Theor Appl Genet 126:2699–2716. https://doi.org/10.1007/s00122-013-2166-x
Su C, Wang W, Gong S, Zuo J, Li S, Xu S (2017) High density linkage map construction and mapping of yield trait QTLs in maize (Zea mays) using the genotyping-by-sequencing (GBS) technology. Front Plant Sci 8:706. https://doi.org/10.3389/fpls.2017.00706
Torkamaneh D, Laroche J, Bastien M, Abed A, Belzile F (2017) Fast-GBS: a new pipeline for the efficient and highly accurate calling of SNPs from genotyping-by-sequencing data. BMC Bioinformatics 18:1–7. https://doi.org/10.1186/s12859-016-1431-9
Tsonev S, Todorovska E, Avramova V, Kolev S, Abu-Mhadi N, Christov N (2009) Genomics assisted improvement of drought tolerance in maize: QTL approaches. Biotechnol Biotechnol Equip 23(4):1410–1413. https://doi.org/10.2478/V10133-009-0004-8
Varshney RK, Nayak SN, May GD, Jackson SA (2009) Next-generation sequencing technologies and their implications for crop genetics and breeding. Trends Biotechnol 27(9):522–530. https://doi.org/10.1016/j.tibtech.2009.05.006
Vierstra RD (2009) The ubiquitin–26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10(6):385–397. https://doi.org/10.1038/nrm2688
Vuylsteke M, Mank R, Antonise R, Bastiaans E, Senior M, Stuber C, Melchinger A, Lübberstedt T, Xia X, Stam P (1999) Two high-density AFLP® linkage maps of Zea mays L.: analysis of distribution of AFLP markers. Theor Appl Genet 99:921–935. https://doi.org/10.1007/s001220051399
Wallace J, Larsson S, Buckler E (2014) Entering the second century of maize quantitative genetics. Heredity 112(1):30–38. https://doi.org/10.1038/hdy.2013.6
Wang B, Hu W, Fang Y, Feng X, Fang J, Zou T, Zheng S, Ming R, Zhang J (2022) Comparative analysis of the MADS-Box genes revealed their potential functions for flower and fruit development in Longan (Dimocarpus longan). Front Plant Sci 12:3259. https://doi.org/10.3389/fpls.2021.813798
Wei T, Simko V (2017). R package “corrplot”: visualization of a correlation matrix.1–5. https://github.com/taiyun/corrplot
Xiu Z, Sun F, Shen Y, Zhang X, Jiang R, Bonnard G, Zhang J, Tan BC (2016) EMPTY PERICARP 16 is required for mitochondrial nad2 intron 4 cis-splicing, complex I assembly and seed development in maize. Plant J 85(4):507–519. https://doi.org/10.1111/tpj.13122
Zeng T, Meng Z, Yue R, Lu S, Li W, Li W, Meng H, Sun Q (2022) Genome wide association analysis for yield related traits in maize. BMC Plant Biol 22(1):449. https://doi.org/10.1186/s12870-022-03812-5
Zhang C, Zhou Z, Yong H, Zhang X, Hao Z, Zhang F, Li M, Zhang D, Li X, Wang Z, Genetics A (2017) Analysis of the genetic architecture of maize ear and grain morphological traits by combined linkage and association mapping. Theor Appl Genet 130:1011–1029. https://doi.org/10.1007/s00122-017-2867-7
Zhang X, Guan Z, Li Z, Liu P, Ma L, Zhang Y, Pan L, He S, Zhang Y, Li P (2020) A combination of linkage mapping and GWAS brings new elements on the genetic basis of yield-related traits in maize across multiple environments. Theor Appl Genet 133:2881–2895. https://doi.org/10.1007/s00122-020-03639-4
Zhang X, Ren Z, Luo B, Zhong H, Ma P, Zhang H, Hu H, Wang Y, Zhang H, Liu D (2022) Genetic architecture of maize yield traits dissected by QTL mapping and GWAS in maize. Crop J 10(2):436–446. https://doi.org/10.1016/j.cj.2021.07.008
Zhou G, Zhu Q, Yang G, Huang J, Cheng S, Yue B, Zhang Z (2015) qEL7. 2 is a pleiotropic QTL for kernel number per row, ear length and ear weight in maize (Zea mays L.). Euphytica 203:429–436. https://doi.org/10.1007/s10681-014-1307-x
Zhou Z, Zhang C, Zhou Y, Hao Z, Wang Z, Zeng X, Di H, Li M, Zhang D, Yong H (2016) Genetic dissection of maize plant architecture with an ultra-high density bin map based on recombinant inbred lines. BMC Genom 17:1–15. https://doi.org/10.1186/s12864-016-2555-z
Zhou Z, Li G, Tan S, Li D, Weiß TM, Wang X, Chen S, Würschum T, Liu W (2020) A QTL atlas for grain yield and its component traits in maize (Zea mays). Plant Breed 139(3):562–574. https://doi.org/10.1111/pbr.12809
Funding
Financial support from the Department of Biotechnology, Government of India, (BT/PR/108910/GBD/27/111/2008 dt.2.6.2008; BT/PR42333/NER/95/1866/2021 dt. 18.2.2022) was acknowledged. The funders had no role in the work design, data collection, and analysis, or decision and preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Communicated by Mian Abdur Rehman Arif.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jadhav, K.P., Karthikeyan, A., Mohanapriya, B. et al. Quantitative trait locus mapping reveals the genomic regions associated with yield-related traits in maize (Zea mays L.). CEREAL RESEARCH COMMUNICATIONS (2024). https://doi.org/10.1007/s42976-024-00510-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42976-024-00510-w