Abstract
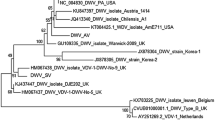
The decline in honey bee colonies in different parts of the world in recent years is due to different reasons, such as agricultural practices, climate changes, the use of chemical insecticides, and pests and diseases. Viral infections are one of the main causes leading to honey bee population declines, which have a major economic impact due to honey production and pollination. To investigate the presence of viruses in bees in southern Brazil, we used a metagenomic approach to sequence adults’ samples of concentrated extracts from Apis mellifera collected in fifteen apiaries of six municipalities in the Rio Grande do Sul state, Brazil, between 2016 and 2017. High-throughput sequencing (HTS) of these samples resulted in the identification of eight previously known viruses (Apis rhabdovirus 1 (ARV-1), Acute bee paralysis virus (ABPV), Aphid lethal paralysis virus (ALPV), Black queen cell virus (BQCV), Bee Macula-like virus (BeeMLV), Deformed wing virus (DWV), Lake Sinai Virus NE (LSV), and Varroa destructor virus 3 (VDV-3)) and a thogotovirus isolate. This thogotovirus shares high amino acid identities in five of the six segments with Varroa orthomyxovirus 1, VOV-1 (98.36 to 99.34% identity). In contrast, segment 4, which codes for the main glycoprotein (GP), has no identity with VOV-1, as observed for the other segments, but shares an amino acid identity of 34–38% with other glycoproteins of viruses from the Orthomyxoviridae family. In addition, the putative thogotovirus GP also shows amino acid identities ranging from 33 to 41% with the major glycoprotein (GP64) of insect viruses of the Baculoviridae family. To our knowledge, this is the second report of a thogotovirus found in bees and given this information, this thogotovirus isolate was tentatively named Apis thogotovirus 1 (ATHOV-1). The detection of multiple viruses in bees is important to better understand the complex interactions between viruses and their hosts. By understanding these interactions, better strategies for managing viral infections in bees and protecting their populations can be developed.






Similar content being viewed by others
Data availability
All data are available under request.
Code availability
Not applicable.
References
Vanbergen AJ, Garratt MP, Vanbergen AJ, et al (2013) Threats to an ecosystem service: pressures on pollinators. Front Ecol Environhttps://doi.org/10.1890/120126
Goulson D, Nicholls E, Botías C, Rotheray EL (2015) Bee declines driven by combined stress from parasites, pesticides, and lack of flowers. Science (80). https://doi.org/10.1126/science.1255957
Klein AM, Vaissière BE, Cane JH et al (2007) Importance of pollinators in changing landscapes for world crops. Proc R Soc B Biol Sci. 274(1608):303–313. https://doi.org/10.1098/rspb.2006.3721
Hung KLJ, Kingston JM, Albrecht M, Holway DA (1870) Kohn JR (2018) The worldwide importance of honey bees as pollinators in natural habitats. Proc R Soc B Biol Sci. 285:20172140. https://doi.org/10.1098/RSPB.2017.2140
De Miranda JR, Bailey L, Ball B V et al (2013) Standard methods for virus research in Apis mellifera. J Apic Res. https://doi.org/10.3896/IBRA.1.52.4.22
Doublet V, Labarussias M, de Miranda JR, Moritz RFA, Paxton RJ (2015) Bees under stress: sublethal doses of a neonicotinoid pesticide and pathogens interact to elevate honey bee mortality across the life cycle. Environ Microbiol. https://doi.org/10.1111/1462-2920.12426
Degrandi-Hoffman G, Chen Y, Watkins Dejong E, Chambers ML, Hidalgo G (2015) Effects of oral exposure to fungicides on honey bee nutrition and virus levels. J Econ Entomol. https://doi.org/10.1093/jee/tov251
Traynor KS, Mondet F, de Miranda JR, et al (2020) Varroa destructor: a complex parasite, crippling honey bees worldwide. Trends Parasitol. https://doi.org/10.1016/j.pt.2020.04.004
Beaurepaire A, Piot N, Doublet V, et al (2020) Diversity and global distribution of viruses of the western honey bee, Apis mellifera. Insects. https://doi.org/10.3390/insects11040239
Matthijs S, Regge N De (2020) Nationwide screening for important bee viruses in Belgian honey bees. Proceedings. https://doi.org/10.3390/proceedings2020050054
Shi M, Lin XD, Tian JH et al (2016) Redefining the invertebrate RNA virosphere. Nature 540(7634):539–543. https://doi.org/10.1038/nature20167
Páez DJ, Fleming-Davies AE (2020) Understanding the evolutionary ecology of host-pathogen interactions provides insights into the outcomes of insect pest biocontrol. Viruses. 12(2):141. https://doi.org/10.3390/v12020141
Kearse M, Moir R, Wilson A et al (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28(12):1647–1649. https://doi.org/10.1093/bioinformatics/bts199
Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30(14):3059–3066
Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Renner S, ed. Syst Biol. 2008;57(5):758–771. https://doi.org/10.1080/10635150802429642
Anisimova M, Gil M, Dufayard JF, Dessimoz C, Gascuel O (2011) Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes. Syst Biol 60(5):685–699
Bin Li K, ClustalW MPI (2003) ClustalW analysis using distributed and parallel computing. Bioinformatics. 19(12):1585–1586. https://doi.org/10.1093/bioinformatics/btg192
Gouet P, Courcelle E, Stuart DI, Métoz F (1999) ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15(4):305–308. https://doi.org/10.1093/bioinformatics/15.4.305
Schwede T, Kopp J, Guex N, Peitsch MC (2003) SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res 31(13):3381–3385. https://doi.org/10.1093/nar/gkg520
Menzel P, Ng KL, Krogh A (2016) Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat Commun 7(1):1–9. https://doi.org/10.1038/ncomms11257
Payne S (2023) Family Polyomaviridae. In: Viruses. Academic Press; 321–325. https://doi.org/10.1016/b978-0-323-90385-1.00043-1
Shi M, Lin XD, Tian JH, et al. Redefining the invertebrate RNA virosphere. Nature. https://doi.org/10.1038/nature20167
Hubálek Z, Rudolf I. Tick-borne viruses in Europe. Parasitol Res. https://doi.org/10.1007/s00436-012-2910-1
Payne S (2017) Family Orthomyxoviridae. In: Viruses. https://doi.org/10.1016/b978-0-12-803109-4.00023-4
Portela A, Jones LD, Nuttall P (1992) Identification of viral structural polypeptides of Thogoto virus (a tick-borne orthomyxo-like virus) and functions associated with the glycoprotein. J Gen Virol 73(11):2823–2830. https://doi.org/10.1099/0022-1317-73-11-2823
Morse MA, Marriott AC, Nuttall PA (1992) The glycoprotein of Thogoto virus (a tick-borne orthomyxo-like virus) is related to the baculovirus glycoprotein GP64. Virology. https://doi.org/10.1016/0042-6822(92)90030-S
Leahy MB, Dessens JT, Weber F, Kochs G, Nuttall PA (1997) The fourth genus in the Orthomyxoviridae: sequence analyses of two Thogoto virus polymerase proteins and comparison with influenza viruses. Virus Res. https://doi.org/10.1016/S0168-1702(97)00072-5
Orthomyxoviridae - negative sense RNA viruses - negative sense RNA viruses (2011) - International Committee on Taxonomy of Viruses (ICTV). Accessed September 4, 2020. https://talk.ictvonline.org/ictv-reports/ictv_9th_report/negative-sense-rna-viruses-2011/w/negrna_viruses/209/orthomyxoviridae
Lung O, Westenberg M, Vlak JM, Zuidema D, Blissard GW (2002) Pseudotyping Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV): F proteins from group II NPVs are functionally analogous to AcMNPV GP64. J Virol 76(11):5729–5736. https://doi.org/10.1128/JVI.76.11.5729-5736.2002
Kuzmin IV, Novella IS, Dietzgen RG, Padhi A, Rupprecht CE (2009) The rhabdoviruses: biodiversity, phylogenetics, and evolution. Infect Genet Evol 9(4):541–553. https://doi.org/10.1016/j.meegid.2009.02.005
Walker PJ, Blasdell KR, Calisher CH et al (2018) ICTV virus taxonomy profile: Rhabdoviridae. J Gen Virol 99(4):447–448. https://doi.org/10.1099/jgv.0.001020
Kuzmin IV, Novella IS, Dietzgen RG, Padhi A, Rupprecht CE (2009) The rhabdoviruses: biodiversity, phylogenetics, and evolution. Infect Genet Evol 9(4):541–553. https://doi.org/10.1016/j.meegid.2009.02.005
Walker PJ, Firth C, Widen SG, et al (2015) Evolution of genome size and complexity in the Rhabdoviridae. PLoS Pathog. https://doi.org/10.1371/journal.ppat.1004664
Valles SM, Chen Y, Firth AE et al (2017) ICTV virus taxonomy profile: Dicistroviridae. J Gen Virol 98(3):355–356. https://doi.org/10.1099/jgv.0.000756
Runckel C, Flenniken ML, Engel JC et al (2011) Temporal analysis of the honey bee microbiome reveals four novel viruses and seasonal prevalence of known viruses, Nosema, and Crithidia. PLoS One. 6(6):e20656. https://doi.org/10.1371/journal.pone.0020656
Daughenbaugh KF, Martin M, Brutscher LM et al (2015) Honey bee infecting Lake Sinai viruses. Viruses 7(6):3285–3309. https://doi.org/10.3390/v7062772
Remnant EJ, Shi M, Buchmann G et al (2017) A diverse range of novel RNA viruses in geographically distinct honey bee populations. J Virol 91(16):158–175. https://doi.org/10.1128/jvi.00158-17
Daughenbaugh KF, Martin M, Brutscher LM, et al (2015) Honey bee infecting Lake Sinai viruses. Viruses. https://doi.org/10.3390/v7062772
Valles SM, Chen Y, Firth AE, et al (2017) ICTV virus taxonomy profile: Iflaviridae. J Gen Virol. https://doi.org/10.1099/jgv.0.000757
Oers MM va. Genomics and biology of lflaviruses. Insect Virol. Published online 2010.
Martelli GP, Sabanadzovic S, Sabanadzovic NAG, Edwards MC, Dreher T (2002) The family Tymoviridae. Arch Virol 147(9):1837–1846. https://doi.org/10.1007/s007050200045
Simmonds P, Adams MJ, Benk M, et al (2017) Consensus statement: Virus taxonomy in the age of metagenomics. Nat Rev Microbiol. https://doi.org/10.1038/nrmicro.2016.177
Unclassified viruses - unclassified viruses - unclassified viruses - ICTV. Accessed March 23, 2021. https://talk.ictvonline.org/ictv-reports/ictv_online_report/unclassified-viruses/w/unclassified-viruses
Backovic M, Jardetzky TS (2009) Class III viral membrane fusion proteins. Curr Opin Struct Biol 19(2):189–196. https://doi.org/10.1016/J.SBI.2009.02.012
Peng R, Zhang S, Cui Y, Shi Y, Gao GF, Qi J (2017) Structures of human-infecting Thogotovirus fusogens support a common ancestor with insect baculovirus. Proc Natl Acad Sci U S A 114(42):E8905–E8912. https://doi.org/10.1073/pnas.1706125114
Bai C, Qi J, Wu Y et al (2019) Postfusion structure of human-infecting Bourbon virus envelope glycoprotein. J Struct Biol 208(2):99–106. https://doi.org/10.1016/j.jsb.2019.08.005
Granberg F, Vicente-Rubiano M, Rubio-Guerri C et al (2013) Metagenomic detection of viral pathogens in Spanish honeybees: co-infection by Aphid lethal paralysis, Israel acute paralysis and Lake Sinai viruses. PLoS One. 8(2):e57459. https://doi.org/10.1371/journal.pone.0057459
Singh R, Levitt AL, Rajotte EG et al (2010) RNA viruses in hymenopteran pollinators: evidence of inter-taxa virus transmission via pollen and potential impact on non-Apis hymenopteran species. PLoS One. 5(12):e14357. https://doi.org/10.1371/journal.pone.0014357
Ravoet J, Maharramov J, Meeus I et al (2013) Comprehensive bee pathogen screening in Belgium reveals Crithidia mellificae as a new contributory factor to winter mortality. PLoS One 8(8):e72443. https://doi.org/10.1371/journal.pone.0072443
Forgách P, Bakonyi T, Tapaszti Z, Nowotny N, Rusvai M (2008) Prevalence of pathogenic bee viruses in Hungarian apiaries: situation before joining the European Union. J Invertebr Pathol 98(2):235–238. https://doi.org/10.1016/j.jip.2007.11.002
McMenamin AJ, Flenniken ML. Recently identified bee viruses and their impact on bee pollinators. Curr Opin Insect Sci. https://doi.org/10.1016/j.cois.2018.02.009
Contreras-Gutiérrez MA, Nunes MRT, Guzman H et al (2017) Sinu virus, a novel and divergent orthomyxovirus related to members of the genus Thogotovirus isolated from mosquitoes in Colombia. Virology 501:166–175. https://doi.org/10.1016/j.virol.2016.11.014
Bolling BG, Weaver SC, Tesh RB, Vasilakis N (2015) Insect-specific virus discovery: significance for the arbovirus community. Viruses 7(9):4911–4928. https://doi.org/10.3390/v7092851
Levin S, Sela N, Chejanovsky N (2016) Two novel viruses associated with the Apis mellifera pathogenic mite Varroa destructor. Sci Rep 6(1):37710. https://doi.org/10.1038/srep37710
Levin S, Sela N, Erez T et al (2019) New viruses from the ectoparasite mite Varroa destructor infesting Apis mellifera and Apis cerana. Viruses 11(2):94. https://doi.org/10.3390/v11020094
Coll JM (1995) The glycoprotein G of rhabdoviruses. Arch Virol 140(5):827–851. https://doi.org/10.1007/BF01314961
Kadlec J, Loureiro S, Abrescia NGA, Stuart DI, Jones IM (2008) The postfusion structure of baculovirus gp64 supports a unified view of viral fusion machines. Nat Struct Mol Biol 15(10):1024–1030. https://doi.org/10.1038/nsmb.1484
Funding
This work was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Grant number 304223/2021–2) and Fundação de apoio à pesquisa do Distrito Federal (FAPDF, Grant number 193001532/2016).
Author information
Authors and Affiliations
Contributions
Bergmann Morais Ribeiro, Lidia Mariana Fiuza, and Daniel Mendes Pereira Ardisson-Araújo designed the study. Leonardo Assis da Silva, Brenda Rabello de Camargo, Bruno Milhomem Pilati Rodrigues, and Diouneia Lisiane Berlitz performed the research. Leonardo Assis da Silva, Daniel Mendes Pereira Ardisson-Araújo, and Bergmann Morais Ribeiro performed the data analysis. Leonardo Assis da Silva, Daniel Mendes Pereira Ardisson-Araújo, and Bergmann Morais Ribeiro wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
All authors consent for publication.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Responsible Editor: María Martha Martorell
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
da Silva, L.A., de Camargo, B.R., Rodrigues, B.M.P. et al. Exploring viral infections in honey bee colonies: insights from a metagenomic study in southern Brazil. Braz J Microbiol 54, 1447–1458 (2023). https://doi.org/10.1007/s42770-023-01078-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-023-01078-z