Abstract
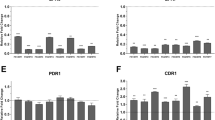
This study aimed to explore the roles of SAP2 and GCN4 in itraconazole (ITR) resistance of C. albicans under different conditions, and their correlations. A total of 20 clinical strains of C. albicans, including 10 ITR resistant strains and 10 sensitive strains, were used. Then, SAP2 sequencing and GCN4 sequencing were performed, and the biofilm formation ability of different C. albicans strains was determined. Finally, real-time quantitative PCR was used to measure the expression of SAP2 and GCN4 in C. albicans under planktonic and biofilm conditions, as well as their correlation was also analyzed. No missense mutations and three synonymous mutation sites, including T276A, G543A, and A675C, were found in SAP2 sequencing. GCN4 sequencing showed one missense mutation site (A106T (T36S)) and six synonymous mutation sites (A147C, C426T, T513C, T576A, G624A and C732T). The biofilm formation ability of drug-resistant C. albicans strains was significantly higher than that of sensitive strains (P < 0.05). Additionally, SAP2 and GCN4 were up-regulated in the ITR-resistant strains, and were both significantly higher in C. albicans under biofilm condition. The mRNA expression levels of SAP2 and GCN4 had significantly positive correlation. The higher expression levels of SAP2 and GCN4 were observed in the ITR-resistant strains of C. albicans under planktonic and biofilm conditions, as well as there was a positive correlation between SAP2 and GCN4 mRNA expression.



Similar content being viewed by others
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Li D, Li T, Bai C et al (2021) A predictive nomogram for mortality of cancer patients with invasive candidiasis: a 10-year study in a cancer center of North China. BMC Infect Dis 21(1):021–05780
Srivastava V, Singla RK, Dubey AK (2018) Emerging Virulence, Drug resistance and future anti-fungal drugs for candida pathogens. Curr Top Med Chem 18(9):759–778
Pinto-Magalhães S, Martins A, Lacerda S et al (2019) Candidemia in a Portuguese tertiary care hospital: analysis of a 2-year period. J Mycol Med 29(4):320–324
Priyadarshini E, Rawat K, Prasad T et al (2018) Antifungal efficacy of Au@ carbon dots nanoconjugates against opportunistic fungal pathogen, Candida albicans. Colloids Surf B Biointerfaces 163:355–361
Pristov KE, Ghannoum MA (2019) Resistance of Candida to azoles and echinocandins worldwide. Clin Microbiol Infect 25(7):792–798. https://doi.org/10.1016/j.cmi.2019.03.028
Peyton LR, Gallagher S, Hashemzadeh M (2015) Triazole antifungals: a review. Drugs Today 51(12):705–718
Lee Y, Puumala E, Robbins N et al (2021) Antifungal drug resistance: molecular mechanisms in Candida albicans and beyond. Chem Rev 121(6):3390–3411. https://doi.org/10.1021/acs.chemrev.0c00199
Vila T, Romo JA, Pierce CG et al (2017) Targeting Candida albicans filamentation for antifungal drug development. Virulence 8(2):150–158. https://doi.org/10.1080/21505594.2016.1197444
Dong S, Shi H, Cao D et al (2016) Novel nanoscale bacteriophage-based single-domain antibodies for the therapy of systemic infection caused by Candida albicans. Sci Rep 6:32256
Aoki W, Kitahara N, Miura N et al (2012) Candida albicans possesses Sap7 as a pepstatin A-insensitive secreted aspartic protease. PLoS One 7(2):27
Dong G, Liu Y, Wu Y et al (2018) Novel non-peptidic small molecule inhibitors of secreted aspartic protease 2 (SAP2) for the treatment of resistant fungal infections. Chem Commun 54(96):13535–13538
Lohse MB, Gulati M, Johnson AD et al (2018) Development and regulation of single- and multi-species Candida albicans biofilms. Nat Rev Microbiol 16(1):19–31
Ponde NO, Lortal L, Ramage G et al (2021) Candida albicans biofilms and polymicrobial interactions. Crit Rev Microbiol 47(1):91–111
Desai JV, Mitchell AP, Andes DR (2014) Fungal biofilms, drug resistance, and recurrent infection. Cold Spring Harb Perspect Med 4(10):a019729
Barros PP, Ribeiro FC, Rossoni RD et al (2016) Influence of Candida krusei and Candida glabrata on Candida albicans gene expression in in vitro biofilms. Arch Oral Biol 64:92–101
Tsui C, Kong EF, Jabra-Rizk MA (2016) Pathogenesis of Candida albicans biofilm. Pathog Dis 74(4):ftw018
Vitális E, Nagy F, Tóth Z et al (2020) Candida biofilm production is associated with higher mortality in patients with candidaemia. Mycoses 63(4):352–360
Monika S, Małgorzata B, Zbigniew O (2017) Contribution of aspartic proteases in Candida virulence. Protease inhibitors against Candida infections. Curr Protein Pept Sci 18(10):1050–62
Hinnebusch AG (2005) Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol 59:407–450
Rana A, Gupta N, Thakur A (2021) Post-transcriptional and translational control of the morphology and virulence in human fungal pathogens. Mol Aspects Med 81(101017):5
Bharadwaj PR, Martins RN (2020) Autophagy modulates Aβ accumulation and formation of aggregates in yeast. Mol Cell Neurosci 104(103466):18
Lee YT, Fang YY, Sun YW et al (2018) THR1 mediates GCN4 and CDC4 to link morphogenesis with nutrient sensing and the stress response in Candida albicans. Int J Mol Med 42(6):3193–3208
Sundaram A, Grant CM (2014) Oxidant-specific regulation of protein synthesis in Candida albicans. Fungal Genet Biol 67:15–23
Sundaram A, Grant CM (2014) A single inhibitory upstream open reading frame (uORF) is sufficient to regulate Candida albicans GCN4 translation in response to amino acid starvation conditions. RNA 20(4):559–567. https://doi.org/10.1261/rna.042267.113
García-Sánchez S, Aubert S, Iraqui I et al (2004) Candida albicans biofilms: a developmental state associated with specific and stable gene expression patterns. Eukaryot Cell 3(2):536–545
Kamthan M, Mukhopadhyay G, Chakraborty N et al (2012) Quantitative proteomics and metabolomics approaches to demonstrate N-acetyl-D-glucosamine inducible amino acid deprivation response as morphological switch in Candida albicans. Fungal Genet Biol 49(5):369–378
Rodriguez DL, Quail MM, Hernday AD et al (2020) Transcriptional circuits regulating developmental processes in Candida albicans. Front Cell Infect Microbiol 10:605711
Candel FJ, Pazos Pacheco C, Ruiz-Camps I et al (2017) Update on management of invasive candidiasis. Rev Esp Quimioter 30(6):397–406
Boija A, Klein IA, Sabari BR et al (2018) Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175(7):1842–1855
Chandra J, Kuhn DM, Mukherjee PK et al (2001) Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J Bacteriol 183(18):5385–5394
Feng W, Yang J, Ma Y et al (2021) The effects of secreted aspartyl proteinase inhibitor ritonavir on azoles-resistant strains of Candida albicans as well as regulatory role of SAP2 and ERG11. Immun Inflamm Dis 9(3):667–680
Liu N, Zhang N, Zhang S et al (2021) Phloretin inhibited the pathogenicity and virulence factors against Candida albicans. Bioengineered 12(1):2420–2431. https://doi.org/10.1080/21655979.2021.1933824
Gulati M, Lohse MB, Ennis CL et al (2018) In vitro culturing and screening of Candida albicans biofilms. Curr Protoc Microbiol 50(1):11
Pereira R, Dos Santos Fontenelle RO, de Brito EHS et al (2021) Biofilm of Candida albicans: formation, regulation and resistance. J Appl Microbiol 131(1):11–22. https://doi.org/10.1111/jam.14949
Chin VK, Lee TY, Rusliza B et al (2016) Dissecting Candida albicans infection from the perspective of C. albicans virulence and omics approaches on host-pathogen interaction: a review. Int J Mol Sci 17(10):1643
Höfs S, Mogavero S, Hube B (2016) Interaction of Candida albicans with host cells: virulence factors, host defense, escape strategies, and the microbiota. J Microbiol 54(3):149–169
Haghighi F, Andriasian L, Tran NC et al (2022) Effect of Cigarette and e-cigarette smoke condensates on Candida albicans biofilm formation and gene expression. Int J Environ Res Public Health 19(8):4626. https://doi.org/10.3390/ijerph19084626
Hu Z, Xia B, Postnikoff SD et al (2018) Ssd1 and Gcn2 suppress global translation efficiency in replicatively aged yeast while their activation extends lifespan. Elife 17(7):35551
Mittal N, Guimaraes JC, Gross T et al (2017) The Gcn4 transcription factor reduces protein synthesis capacity and extends yeast lifespan. Nat Commun 8(1):017–00539
Shen ZJ, Postnikoff S, Tyler JK (2019) Is Gcn4-induced autophagy the ultimate downstream mechanism by which hormesis extends yeast replicative lifespan? Curr Genet 65(3):717–720
Feng W, Yang J, Pan Y et al (2016) The correlation of virulence, pathogenicity, and itraconazole resistance with SAP activity in Candida albicans strains. Can J Microbiol 62(2):173–178. https://doi.org/10.1139/cjm-2015-0457
Funding
This study was supported by the General Project of the National Natural Science Foundation of China (Project number: 82072262), and Research and Development Key Projects of Shanxi Province (Project number: 201903D321123), Scientific and Technological Activities funding program for Overseas Students of Shanxi Province (Project number:20210030), and Research Project Supported by Shanxi Scholarship Council of China (Project number: 2020–190).
Author information
Authors and Affiliations
Contributions
Wenli Feng and Jing Yang designed the research. Wenli Feng, Jing Yang, Yan Ma, Luwen Zhang, and Rong Yin did the experiment and obtained the data. Zusha Qiao, Ying Ji, and Yong’an Zhou analyzed and explained the data. Wenli Feng and Jing Yang drafted and revised the manuscript. All authors have read and approved the final version.
Corresponding authors
Ethics declarations
Ethics approval
This study was approved by the Ethics Committee of the Second Hospital of Shanxi Medical University (approval no. (2019)YX no. 073).
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Responsible Editor: Rosana Puccia
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Feng, W., Yang, J., Ma, Y. et al. Relationships between Secreted Aspartyl Proteinase 2 and General Control Nonderepressible 4 gene in the Candida albicans resistant to itraconazole under planktonic and biofilm conditions. Braz J Microbiol 54, 619–627 (2023). https://doi.org/10.1007/s42770-023-00961-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-023-00961-z