Abstract
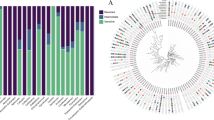
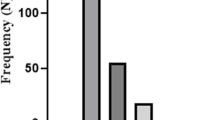
Infection with P. aeruginosa, one of the most relevant opportunistic pathogens in hospital-acquired infections, can lead to high mortality due to its low antibiotic susceptibility to limited choices of antibiotics. Polymyxin as last-resort antibiotics is used in the treatment of systemic infections caused by multidrug-resistant P. aeruginosa strains, so studying the emergence of polymyxin-resistant was a must. The present study was designed to define genomic differences between paired polymyxin-susceptible and polymyxin-resistant P. aeruginosa strains and established polymyxin resistance mechanisms, and common chromosomal mutations that may confer polymyxin resistance were characterized. A total of 116 CRPA clinical isolates from patients were collected from three tertiary care hospitals in China during 2017–2021. Our study found that polymyxin B resistance represented 3.45% of the isolated carbapenem-resistant P. aeruginosa (CRPA). No polymyxin-resistant isolates were positive for mcr (1–8 and 10) gene and efflux mechanisms. Key genetic variations identified in polymyxin-resistant isolates involved missense mutations in parR, parS, pmrB, pmrA, and phoP. The waaL and PA5005 substitutions related to LPS synthesis were detected in the highest levels of resistant strain (R1). The missense mutations H398R in ParS (4/4), Y345H in PmrB (4/4), and L71R in PmrA (3/4) were the predominant. Results of the PCR further confirmed that mutation of pmrA, pmrB, and phoP individually or simultaneously did affect the expression level of resistant populations and can directly increase the expression of arnBCADTEF operon to contribute to polymyxin resistance. In addition, we reported 3 novel mutations in PA1945 (2129872_A < G, 2130270_A < C, 2130272_T < G) that may confer polymyxin resistance in P. aeruginosa. Our findings enriched the spectrum of chromosomal mutations, highlighted the complexity at the molecular level, and multifaceted interplay mechanisms underlying polymyxin resistance in P. aeruginosa.





Similar content being viewed by others
Data availability
Some or all data, models, or code generated or used during the study are available from the corresponding author by request.
References
Zhu Y, Czauderna T, Zhao J, Klapperstueck M, Maifiah MHM, Han M-L, Lu J, Sommer B, Velkov T, Lithgow T, Song J, Schreiber F, Li J (2018) Genome-scale metabolic modeling of responses to polymyxins in Pseudomonas aeruginosa. Gigascience 7(4):giy021. https://doi.org/10.1093/gigascience/giy021
Horcajada JP, Montero M, Oliver A, Sorlí L, Luque S, Gómez-Zorrilla S, Benito N, Grau S (2019) Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin Microbiol Rev 32(4):e00031-e119. https://doi.org/10.1128/CMR.00031-19
Lee J-Y, Na IY, Park YK, Ko KS (2014) Genomic variations between colistin-susceptible and -resistant Pseudomonas aeruginosa clinical isolates and their effects on colistin resistance. J Antimicrob Chemother 69(5):1248–1256. https://doi.org/10.1093/jac/dkt531
Jeannot K, Bolard A, Plésiat P (2017) Resistance to polymyxins in Gram-negative organisms. Int J Antimicrob Agents 49(5):526–535. https://doi.org/10.1016/j.ijantimicag
Xiao C, Zhu Y, Yang Z, Shi D, Ni Y, Hua Li, Li J (2022) Prevalence and molecular characteristics of polymyxin-resistant Pseudomonas aeruginosa in a Chinese tertiary teaching hospital. Antibiotics (Basel) 11(6):799. https://doi.org/10.3390/antibiotics11060799
Landman D, Bratu S, Alam M, Quale J (2005) Citywide emergence of Pseudomonas aeruginosa strains with reduced susceptibility to polymyxin B. J Antimicrob Chemother 55:954–957. https://doi.org/10.1093/jac/dki153
Lee JY, Song JH, Ko KS (2011) Identification of nonclonal Pseudomonas aeruginosa isolates with reduced colistin susceptibility in Korea. Microb Drug Resist 17:299–304. https://doi.org/10.1089/mdr.2010.0145
Orsi TD, Neto LVP, Martins RCR, Levin AS, Costa SF (2019) Polymyxin-resistant Pseudomonas aeruginosa assigned as ST245: first report in an intensive care unit in São Paulo, Brazil. J Glob Antimicrob Resist 16:147–149. https://doi.org/10.1016/j.jgar.2018.12.021
CianciulliSesso A, Lilić B, Amman F, Wolfinger MT, Sonnleitner E, Bläsi U (2021) Gene expression profiling of Pseudomonas aeruginosa upon exposure to colistin and tobramycin. Front Microbiol 12:626715. https://doi.org/10.3389/fmicb.2021.626715
Puja H, Bolard A, Noguès A, Plésiat P, Jeannot K (2020) The efflux pump MexXY/OprM contributes to the tolerance and acquired resistance of Pseudomonas aeruginosa to colistin. Antimicrob Agents Chemother 64(4):1–11. https://doi.org/10.1128/AAC.02033-19
Bell A, Bains M, Hancock RE (1991) Pseudomonas aeruginosa outer membrane protein OprH: expression from the cloned gene and function in EDTA and gentamicin resistance. J Bacteriol 173(21):6657–6664. https://doi.org/10.1128/jb.173.21.6657-6664.1991
Lin J, Xu C, Fang R, Cao J, Zhang X, Zhao Y, Dong G, Sun Y, Zhou T (2019) Resistance and heteroresistance to colistin in Pseudomonas aeruginosa isolates from Wenzhou, China. Antimicrob Agents Chemother 63(10):e00556-19. https://doi.org/10.1128/AAC.00556-19
Nang SC, Li J, Velkov T (2019) The rise and spread of mcr plasmid-mediated polymyxin resistance. Crit Rev Microbiol 45(2):131–161. https://doi.org/10.1080/1040841X.2018.1492902
Poole K, Lau C-F, Gilmou C, Hao Y, Lam JS (2015) Polymyxin susceptibility in Pseudomonas aeruginosa linked to the MexXY-OprM multidrug efflux system. Antimicrob Agents Chemother 59(12):7276–7289. https://doi.org/10.1128/AAC.01785-15
Yang B, Liu C, Pan X, Weixin Fu, Fan Z, Jin Y, Bai F, Cheng Z, Weihui Wu (2021) Identification of novel PhoP-PhoQ regulated genes that contribute to polymyxin B tolerance in Pseudomonas aeruginosa. Microorganisms 9(2):344. https://doi.org/10.3390/microorganisms9020344
Yang Q, Pogue JM, Li Z, Nation RL, Kaye KS, Li J (2020) Agents of last resort: an update on polymyxin resistance. Infect Dis Clin North Am Dec 34(4):723–750. https://doi.org/10.1016/j.idc.2020.08.003
Huszczynski SM, Lam JS, Khursigara CM (2019) The role of Pseudomonas aeruginosa lipopolysaccharide in bacterial pathogenesis and physiology. Pathogens 9(1):6. https://doi.org/10.3390/pathogens9010006
Sciuto AL, Imperi F (2018) Aminoarabinosylation of lipid A is critical for the development of colistin resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 62(3):e01820-e1917. https://doi.org/10.1128/AAC.01820-17
Pedersen MG, Olesen HV, Jensen-Fangel S, Nørskov-Lauritsen N, Wang M (2018) Colistin resistance in Pseudomonas aeruginosa and Achromobacter spp. cultured from Danish cystic fibrosis patients is not related to plasmid-mediated expression of mcr-1. J Cyst Fibros 17(2):e22–e23. https://doi.org/10.1016/j.jcf.2017.12.001
Lee JY, Ko KS (2014) Mutations and expression of PmrAB and PhoPQ related with colistin resistance in Pseudomonas aeruginosa clinical isolates. Diagn Microbiol Infect Dis 78(3):271–276. https://doi.org/10.1016/j.diagmicrobio.2013.11.027
Owusu-Anim D, Kwon DH (2012) Differential role of two-component regulatory systems (phoPQ and pmrAB) in polymyxin B susceptibility of Pseudomonas aeruginosa. Adv Microbiol 2(1):1–10. https://doi.org/10.4236/aim.2012.21005
Lee JY, Chung ES, Na IY, Kim H, Shin D, Ko KS (2014) Development of colistin resistance in pmrA-, phoP-, parR- and cprR-inactivated mutants of Pseudomonas aeruginosa. J AntimicrobChemother 69(11):2966–2971. https://doi.org/10.1093/jac/dku238
Jochumsen N, Marvig RL, Damkiær S, Jensen RL, Paulander W, Molin S, Jelsbak L, Folkesson A (2016) The evolution of antimicrobial peptide resistance in Pseudomonas aeruginosa is shaped by strong epistatic interactions. Nat Commun 7:1–10. https://doi.org/10.1038/ncomms13002
Lee JY, Park YK, Chung ES, Na IY, Ko KS (2016) Evolved resistance to colistin and its loss due to genetic reversion in Pseudomonas aeruginosa. Sci Rep 6:1–12. https://doi.org/10.1038/srep25543
Acknowledgements
This work was supported by grants from the Special Foundation for National Science and Technology Basic Research Program of China (2019FY101200, 2019FY101209), the Yunnan Health Training Project of High Level Talents (Grant No: H-2018075), and the Yunnan High-Level Personnel Training Support Plan (Grant No: YNWR-QNBJ-2020-261).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Responsible Editor: Luis Augusto Nero
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liang, Y., Li, J., Xu, Y. et al. Genomic variations in polymyxin-resistant Pseudomonas aeruginosa clinical isolates and their effects on polymyxin resistance. Braz J Microbiol 54, 655–664 (2023). https://doi.org/10.1007/s42770-023-00933-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-023-00933-3