Abstract
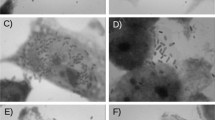
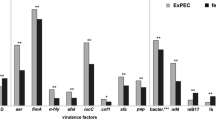
Extraintestinal pathogenic Escherichia coli (ExPEC) is the leading cause of urinary tract infection worldwide and a critical bloodstream infection agent. There are more than 50 virulence factors (VFs) related to ExPEC pathogenesis; however, many strains isolated from extraintestinal infections are devoid of these factors. Since opportunistic infections may occur in immunocompromised patients, E. coli strains that lack recognized VFs are considered opportunist, and their virulence potential is neglected. We assessed eleven E. coli strains isolated from bloodstream infections and devoid of the most common ExPEC VFs to understand their pathogenic potential. The strains were evaluated according to their capacity to interact in vitro with human eukaryotic cell lineages (Caco-2, T24, HEK293T, and A549 cells), produce type 1 fimbriae and biofilm in diverse media, resist to human sera, and be lethal to Galleria mellonella. One strain displaying all phenotypic traits was sequenced and evaluated. Ten strains adhered to Caco-2 (colon), eight to T24 (bladder), five to HEK-293 T (kidney), and four to A549 (lung) cells. Eight strains produced type 1 fimbriae, ten adhered to abiotic surfaces, nine were serum resistant, and seven were virulent in the G. mellonella model. Six of the eleven E. coli strains displayed traits compatible with pathogens, five of which were isolated from an immune-competent host. The genome of the EC175 strain, isolated from a patient with urosepsis, reveals that the strain belonged to ST504-A, and serotype O11:H11; harbors thirteen VFs genes, including genes encoding UpaG and yersiniabactin as the only ExPEC VFs identified. Together, our results suggest that the ExPEC pathotype includes pathogens from phylogroups A and B1, which harbor VFs that remain to be uncovered.





Similar content being viewed by others
Data availability
The Whole Genome Shotgun project of E. coli EC175 has been deposited at DDBJ/ENA/GenBank under the accession JALLIP000000000. The version described in this article is version JALLIP010000000.
References
Leimbach A, Hacker J, Dobrindt U (2013) E. coli as an all-rounder: the thin line between commensalism and pathogenicity. Curr Top Microbiol Immunol 358:3–32. https://doi.org/10.1007/82_2012_303
Martinson JNv, Walk ST (2020) Escherichia coli residency in the gut of healthy human adults. EcoSal Plus 9(1). https://doi.org/10.1128/ecosalplus.esp-0003-2020
Johnson JR, Russo TA (2018) Molecular epidemiology of extraintestinal pathogenic Escherichia coli. EcoSal Plus 8(1). https://doi.org/10.1128/ecosalplus.ESP-0004-2017
Ewers C, Li G, Wilking H et al (2007) Avian pathogenic, uropathogenic, and newborn meningitis-causing Escherichia coli: how closely related are they? Int J Med Microbiol 297(3):163–176. https://doi.org/10.1016/j.ijmm.2007.01.003
Santos ACM, Santos FF, Silva RM, Gomes TAT (2020) Diversity of hybrid- and hetero-pathogenic Escherichia coli and their potential implication in more severe diseases. Front Cell Infect Microbiol 10:339. https://doi.org/10.3389/FCIMB.2020.00339
Mellata M (2013) Human and avian extraintestinal pathogenic Escherichia coli: infections, zoonotic risks, and antibiotic resistance trends. Foodborne Pathog Dis 10(11):916–932. https://doi.org/10.1089/fpd.2013.1533
Manges AR, Geum HM, Guo A, Edens TJ, Fibke CD, Pitout JDD (2019) Global extraintestinal pathogenic Escherichia coli (ExPEC) lineages. Clin Microbiol Rev 32(3). https://doi.org/10.1128/CMR.00135-18
Messika J, Magdoud F, Clermont O et al (2012) Pathophysiology of Escherichia coli ventilator-associated pneumonia: implication of highly virulent extraintestinal pathogenic strains. Intensive Care Med 38(12):2007–2016. https://doi.org/10.1007/s00134-012-2699-5
Nunes PHS, Valiatti TB, de Santos ACM et al (2022) Evaluation of the pathogenic potential of Escherichia coli strains isolated from eye infections. Microorganisms 10(6):1084. https://doi.org/10.3390/microorganisms10061084
Santos ACM, Fuga B, Esposito F et al (2021) Unveiling the virulent genotype and unusual biochemical behavior of Escherichia coli ST59. Appl Environ Microbiol 87(16):e0074321. https://doi.org/10.1128/AEM.00743-21
Köhler CD, Dobrindt U (2011) What defines extraintestinal pathogenic Escherichia coli? Int J Med Microbiol 301(8):642–647. https://doi.org/10.1016/J.IJMM.2011.09.006
Freire CA, Santos ACM, Pignatari AC, Silva RM, Elias WP (2020) Serine protease autotransporters of Enterobacteriaceae (SPATEs) are largely distributed among Escherichia coli isolated from the bloodstream. Braz J Microbiol 51(2):447–454. https://doi.org/10.1007/s42770-020-00224-1
Santos ACM, Zidko ACM, Pignatari AC, Silva RM (2013) Assessing the diversity of the virulence potential of Escherichia coli isolated from bacteremia in São Paulo, Brazil. Braz J Med Biol Res 46(11):968–973. https://doi.org/10.1590/1414-431X20133184
Santos ACM. Virulence potential and antimicrobial susceptibility of Escherichia coli strains isolated from bacteremia. Their relationship with immunological competence of the host, and the origin of infection. PhD Thesis. Universidade Federal de São Paulo; 2013. Accessed June 19, 2019. http://repositorio.unifesp.br/handle/11600/22975
Santos ACM, Silva RM, Valiatti TB et al (2020) Virulence potential of a multidrug-resistant Escherichia coli strain belonging to the emerging clonal group ST101-B1 isolated from bloodstream infection. Microorganisms 8(6):827. https://doi.org/10.3390/microorganisms8060827
Clermont O, Christenson JK, Denamur E, Gordon DM (2013) The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep 5(1):58–65. https://doi.org/10.1111/1758-2229.12019
Dozois CM, Clément S, Desautels C, Oswald E, Fairbrother JM (2006) Expression of P, S, and F1C adhesins by cytotoxic necrotizing factor 1-producing Escherichia coli from septicemic and diarrheic pigs. FEMS Microbiol Lett 152(2):307–312. https://doi.org/10.1111/j.1574-6968.1997.tb10444.x
Valiatti TB, Santos FF, Santos ACM et al (2020) Genetic and virulence characteristics of a hybrid atypical enteropathogenic and uropathogenic Escherichia coli (aEPEC/UPEC) strain. Front Cell Infect Microbiol 10:492. https://doi.org/10.3389/fcimb.2020.00492
Malberg Tetzschner AM, Johnson JR, Johnston BD, Lund O, Scheutz F (2020) In silico genotyping of Escherichia coli isolates for extraintestinal virulence genes by use of whole-genome sequencing data. J Clin Microbiol. https://doi.org/10.1128/JCM.01269-20
Joensen KG, Scheutz F, Lund O et al (2014) Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol 52(5):1501–1510. https://doi.org/10.1128/JCM.03617-13
Camacho C, Coulouris G, Avagyan V et al (2009) BLAST+: architecture and applications. BMC Bioinformatics 10. https://doi.org/10.1186/1471-2105-10-421
Zankari E, Allesøe R, Joensen KG, Cavaco LM, Lund O, Aarestrup FM (2017) PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J Antimicrob Chemother 72(10):2764–2768. https://doi.org/10.1093/jac/dkx217
Bortolaia V, Kaas RS, Ruppe E et al (2020) ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother 75(12):3491–3500. https://doi.org/10.1093/JAC/DKAA345
Carattoli A, Zankari E, García-Fernández A et al (2014) In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58(7):3895–3903. https://doi.org/10.1128/AAC.02412-14
Roer L, Johannesen TB, Hansen F et al (2018) CHTyper, a web tool for subtyping of extraintestinal pathogenic Escherichia coli based on the fumC and fimH alleles. J Clin Microbiol 56(4):63–81. https://doi.org/10.1128/JCM.00063-18
Wirth T, Falush D, Lan R et al (2006) Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60(5):1136–1151. https://doi.org/10.1111/j.1365-2958.2006.05172.x
Liu B, Zheng D, Jin Q, Chen L, Yang J (2018) VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res 47:687–692. https://doi.org/10.1093/nar/gky1080
Wattam AR, Davis JJ, Assaf R et al (2017) Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res 45(D1):D535–D542. https://doi.org/10.1093/nar/gkw1017
Letunic I, Bork P (2021) Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. https://doi.org/10.1093/nar/gkab301
Denamur E, Clermont O, Bonacorsi S, Gordon D (2020) The population genetics of pathogenic Escherichia coli. Nat Rev Microbiol 19(1):1–18. https://doi.org/10.1038/s41579-020-0416-x
Lüthje P, Brauner A (2014) Virulence factors of uropathogenic E. coli and their interaction with the host. Adv Microb Physiol 65:337–372. https://doi.org/10.1016/bs.ampbs.2014.08.006
Müller CM, Åberg A, Straseviçiene J, Emody L, Uhlin BE, Balsalobre C (2009) Type 1 fimbriae, a colonization factor of uropathogenic Escherichia coli, are controlled by the metabolic sensor CRP-cAMP. PLoS Pathog 5(2). https://doi.org/10.1371/journal.ppat.1000303
Johnson JR, Weissman SJ, Stell AL, Trintchina E, Dykhuizen DE, Sokurenko Ev (2001) Clonal and pathotypic analysis of archetypal Escherichia coli cystitis isolate NU14. J Infect Dis 184(12):1556–1565. https://doi.org/10.1086/323891
Sokurenko Ev, Chesnokova V, Dykhuizen DE et al (1998) Pathogenic adaptation of Escherichia coli by natural variation of the FimH adhesin. Proc Natl Acad Sci U S A 95(15):8922–8926. https://doi.org/10.1073/PNAS.95.15.8922
Mydock-McGrane L, Cusumano Z, Han Z et al (2016) Antivirulence C-mannosides as antibiotic-sparing, oral therapeutics for urinary tract infections. J Med Chem 59(20):9390–9408. https://doi.org/10.1021/acs.jmedchem.6b00948
Neugent ML, Hulyalkar NV, Nguyen VH, Zimmern PE, De Nisco NJ (2020) Advances in understanding the human urinary microbiome and its potential role in urinary tract infection. MBio 11(2). https://doi.org/10.1128/mBio.00218-20
Eto DS, Jones TA, Sundsbak JL, Mulvey MA (2007) Integrin-mediated host cell invasion by type 1–piliated uropathogenic Escherichia coli. PLoS Pathog 3(7):e100. https://doi.org/10.1371/JOURNAL.PPAT.0030100
Connell H, Agace W, Klemm P, Schembri M, Mårild S, Svanborg C (1996) Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc Natl Acad Sci U S A 93(18):9827–9832. https://doi.org/10.1073/pnas.93.18.9827
Watts RE, Hancock V, Ong CLY et al (2010) Escherichia coli isolates causing asymptomatic bacteriuria in catheterized and noncatheterized individuals possess similar virulence properties. J Clin Microbiol 48(7):2449–2458. https://doi.org/10.1128/JCM.01611-09
Avalos Vizcarra I, Hosseini V, Kollmannsberger P et al (2015) How type 1 fimbriae help Escherichia coli to evade extracellular antibiotics. Sci Rep 2016(6):1–13. https://doi.org/10.1038/srep18109
Kim KS (2016) Human meningitis-associated Escherichia coli. EcoSal Plus 7(1). https://doi.org/10.1128/ECOSALPLUS.ESP-0015-2015
Bessaiah H, Anamalé C, Sung J, Dozois CM (2021) What flips the switch? Signals and stress regulating extraintestinal pathogenic Escherichia coli type 1 fimbriae (pili). Microorganisms 10(5):5. https://doi.org/10.3390/MICROORGANISMS10010005
Miyazaki J, Ba-Thein W, Kumao T et al (2002) Type 1, P and S fimbriae, and afimbrial adhesin I are not essential for uropathogenic Escherichia coli to adhere to and invade bladder epithelial cells. FEMS Immunol Med Microbiol 33(1):23–26
Korea CG, Badouraly R, Prevost MC, Ghigo JM, Beloin C (2010) Escherichia coli K-12 possesses multiple cryptic but functional chaperone-usher fimbriae with distinct surface specificities. Environ Microbiol 12(7):1957–1977. https://doi.org/10.1111/j.1462-2920.2010.02202.x
Snyder JA, Haugen BJ, Lockatell CV et al (2005) Coordinate expression of fimbriae in uropathogenic Escherichia coli. Infect Immun 73(11):7588–7596. https://doi.org/10.1128/IAI.73.11.7588-7596.2005
Saldaña Z, De la Cruz MA, Carrillo-Casas EM et al (2014) Production of the Escherichia coli common pilus by uropathogenic E. coli is associated with adherence to HeLa and HTB-4 cells and invasion of mouse bladder urothelium. PLoS One 9(7):e101200. https://doi.org/10.1371/journal.pone.0101200
Rendón MA, Saldaña Z, Erdem AL et al (2007) Commensal and pathogenic Escherichia coli use a common pilus adherence factor for epithelial cell colonization. Proc Natl Acad Sci U S A 104(25):10637–10642. https://doi.org/10.1073/PNAS.0704104104
Rossez Y, Holmes A, Lodberg-Pedersen H et al (2014) Escherichia coli common pilus (ECP) targets arabinosyl residues in plant cell walls to mediate adhesion to fresh produce plants. J Biol Chem 289(49):34349–34365. https://doi.org/10.1074/JBC.M114.587717
Lehti TA, Bauchart P, Heikkinen J et al (2010) Mat fimbriae promote biofilm formation by meningitis-associated Escherichia coli. Microbiology (N Y) 156(8):2408–2417. https://doi.org/10.1099/MIC.0.039610-0
Pouttu R, Westerlund-Wikström B, Lång H et al (2001) matB, a common fimbrillin gene of Escherichia coli, expressed in a genetically conserved, virulent clonal group. J Bacteriol 183(16):4727–4736. https://doi.org/10.1128/JB.183.16.4727-4736.2001
Day CJ, Lo AW, Hartley-Tassell LE et al (2021) Discovery of bacterial fimbria–glycan interactions using whole-cell recombinant Escherichia coli expression. mBio 12(1):1–10. https://doi.org/10.1128/MBIO.03664-20/SUPPL_FILE/MBIO.03664-20-ST001.DOCX
Novais Â, Pires J, Ferreira H et al (2012) Characterization of globally spread Escherichia coli ST131 isolates (1991 to 2010). Antimicrob Agents Chemother 56(7):3973–3976. https://doi.org/10.1128/AAC.00475-12
Ponnusamy P, Natarajan V, Sevanan M (2012) In vitro biofilm formation by uropathogenic Escherichia coli and their antimicrobial susceptibility pattern. Asian Pac J Trop Med 5(3):210–213. https://doi.org/10.1016/S1995-7645(12)60026-1
Agarwal J, Mishra B, Srivastava S, Srivastava R (2013) Genotypic characteristics and biofilm formation among Escherichia coli isolates from Indian women with acute cystitis. Trans R Soc Trop Med Hyg 107(3):183–187. https://doi.org/10.1093/trstmh/trs090
Tapiainen T, Hanni AM, Salo J, Ikäheimo I, Uhari M (2014) Escherichia coli biofilm formation and recurrences of urinary tract infections in children. Eur J Clin Microbiol Infect Dis 33(1):111–115. https://doi.org/10.1007/s10096-013-1935-4
Flament-Simon SC, Nicolas-Chanoine MH, García V et al (2020) Clonal structure virulence factor-encoding genes and antibiotic resistance of Escherichia coli causing urinary tract infections and other extraintestinal infections in humans in Spain and France during 2016. Antibiotics 9(4):161. https://doi.org/10.3390/ANTIBIOTICS9040161
Pont S, Fraikin N, Caspar Y, van Melderen L, Attrée I, Cretin F (2020) Bacterial behavior in human blood reveals complement evaders with some persister-like features. PLoS Pathog. 16(12). https://doi.org/10.1371/JOURNAL.PPAT.1008893
Miajlovic H, Smith SG (2014) Bacterial self-defence: how Escherichia coli evades serum killing. FEMS Microbiol Lett 354(1):1–9. https://doi.org/10.1111/1574-6968.12419
Johnson JR (1991) Virulence factors in Escherichia coli urinary tract infection. Clin Microbiol Rev 4(1):80–128. https://doi.org/10.1128/CMR.4.1.80
Biran D, Rosenshine I, Ron EZ (2020) Escherichia coli O-antigen capsule (group 4) is essential for serum resistance. Res Microbiol 171(2):99–101. https://doi.org/10.1016/J.RESMIC.2019.12.002
Dutra IL, Araújo LG, Assunção RG et al (2020) Pic-producing Escherichia coli induces high production of proinflammatory mediators by the host leading to death by sepsis. Int J Mol Sci 21(6):2068. https://doi.org/10.3390/ijms21062068
Freire CA, Silva RM, Ruiz RC et al (2022) Secreted autotransporter toxin (Sat) mediates innate immune system evasion. Front Immunol 13:433. https://doi.org/10.3389/FIMMU.2022.844878/BIBTEX
Weiser JN, Gotschlich EC (1991) Outer membrane protein A (OmpA) contributes to serum resistance and pathogenicity of Escherichia coli K-1. Infect Immun 59(7):2252–2258. https://doi.org/10.1128/IAI.59.7.2252-2258.1991
Coggon CF, Jiang A, Goh KGK, Henderson IR, Schembri MA, Wells TJ (2018) A novel method of serum resistance by Escherichia coli that causes urosepsis. mBio. 9(3). https://doi.org/10.1128/MBIO.00920-18
Liu J, Huang L, Luo M, Xia X (2019) Bacterial translocation in acute pancreatitis. Crit Rev Microbiol 45(5–6):539–547. https://doi.org/10.1080/1040841X.2019.1621795
Ma D, Jiang P, Jiang Y, Li H, Zhang D (2021) Effects of lipid peroxidation-mediated ferroptosis on severe acute pancreatitis-induced intestinal barrier injury and bacterial translocation. Oxid Med Cell Longev. 2021. https://doi.org/10.1155/2021/6644576
Ciesielczuk H, Betts J, Phee L et al (2015) Comparative virulence of urinary and bloodstream isolates of extra-intestinal pathogenic Escherichia coli in a Galleria mellonella model. Virulence 6(2):145–151. https://doi.org/10.4161/21505594.2014.988095
Williamson DA, Mills G, Johnson JR, Porter S, Wiles S (2014) In vivo correlates of molecularly inferred virulence among extraintestinal pathogenic Escherichia coli (ExPEC) in the wax moth Galleria mellonella model system. Virulence 5(3):388–393. https://doi.org/10.4161/viru.27912
Heitmueller M, Billion A, Dobrindt U, Vilcinskas A, Mukherjee K (2017) Epigenetic mechanisms regulate innate immunity against uropathogenic and commensal-like Escherichia coli in the surrogate insect model Galleria mellonella. Infect Immun 85(10):e00336–17. https://doi.org/10.1128/IAI.00336-17
Alghoribi MF, Gibreel TM, Dodgson AR, Beatson SA, Upton M (2014) Galleria mellonella infection model demonstrates high lethality of ST69 and ST127 uropathogenic E. coli. PLoS One 9(7):e101547. https://doi.org/10.1371/journal.pone.0101547
Pereira TC, Barros PP de, de Oliveira Fugisaki LR et al (2018) Recent advances in the use of Galleria mellonella model to study immune responses against human pathogens. J Fungi 4(4):128. https://doi.org/10.3390/JOF4040128
Valle J, Mabbett AN, Ulett GC et al (2008) UpaG, a new member of the trimeric autotransporter family of adhesins in uropathogenic Escherichia coli. J Bacteriol 190(12):4147–4161. https://doi.org/10.1128/JB.00122-08
William O’Hara R, Jenks PJ, Emery M, Upton M (2019) Rapid detection of extra-intestinal pathogenic Escherichia coli multi-locus sequence type 127 using a specific PCR assay. J Med Microbiol 68(2):188–196. https://doi.org/10.1099/JMM.0.000902
Totsika M, Wells TJ, Beloin C et al (2012) Molecular characterization of the EhaG and UpaG trimeric autotransporter proteins from pathogenic Escherichia coli. Appl Environ Microbiol 78(7):2179–2189. https://doi.org/10.1128/AEM.06680-11
Sikdar R, Bernsteina HD (2019) Sequential translocation of polypeptides across the bacterial outer membrane through the trimeric autotransporter pathway. MBio 10(5). https://doi.org/10.1128/MBIO.01973-19
Carniel E, Guilvout I, Prentice M (1996) Characterization of a large chromosomal “high-pathogenicity island” in biotype 1B Yersinia enterocolitica. J Bacteriol 178(23):6743. https://doi.org/10.1128/JB.178.23.6743-6751.1996
Buchrieser C, Prentice M, Carniel E (1998) The 102-kilobase unstable region of Yersinia pestis comprises a high- pathogenicity island linked to a pigmentation segment which undergoes internal rearrangement. J Bacteriol 180(9):2321–2329. https://doi.org/10.1128/JB.180.9.2321-2329.1998
Galardini M, Clermont O, Baron A et al (2020) Major role of iron uptake systems in the intrinsic extra-intestinal virulence of the genus Escherichia revealed by a genome-wide association study. PLoS Genet 16(10):e1009065. https://doi.org/10.1371/journal.pgen.1009065
Acknowledgements
We are grateful to Prof. Dr. Erika Suzuki and Prof. Dr. Nilton Lincopan for providing the A549 cell lineage, and the G. mellonella larvae, respectively. EC175 genome was sequenced using MicrobesNG (Birmingham Research Park, Birmingham, UK) sequencing service.
Funding
This study was supported by the National Council for Science and Technological Development (CNPq) [304760/2015–3 and 311283/2020–9 to TATG] and by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) [2017/14821-7 and 2018/17353–7 to TATG]; ACMS received PNPD fellowship [88882.306532/2018–01] from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (Capes) under financial code 001; JFSN received scholarship from FAPESP [2019/21685–8], and LOT from CNPq [168193/2018–3].
Author information
Authors and Affiliations
Contributions
Conceptualization: ACMS, RMS, and TATG. Formal analysis: ACMS. Funding acquisition: TATG. Investigation: ACMS, JFSN, LOT, and RFTR. Validation: ACMS, JFSN, and LOT. Project administration: ACMS. Resources: RMS, TATG. Supervision: ACMS, TATG. Visualization: ACMS. Writing — original draft: ACMS. Writing — review and editing: ACMS, JFSN, RMS, and TATG.
Corresponding authors
Ethics declarations
Ethical approval
This research was done with approval of the Research Ethics Committee of the Federal University of São Paulo—UNIFESP/São Paulo Hospital (CEP N 7140160317 from April 2017).
Conflict of interest
The authors declare no competing interests.
Additional information
Responsible Editor: Luis Nero
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Santos, A.C.M., Santos-Neto, J.F., Trovão, L.O. et al. Characterization of unconventional pathogenic Escherichia coli isolated from bloodstream infection: virulence beyond the opportunism. Braz J Microbiol 54, 15–28 (2023). https://doi.org/10.1007/s42770-022-00884-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-022-00884-1