Abstract
Purpose
Nasopharyngeal carcinoma (NPC), an epithelial-originated malignant tumor, has a special geographic distribution. However, the etiology of NPC has not been examined in detail. Increasing pieces of evidence indicate that the microbiome may contribute to head and neck squamous cell carcinoma. Until now, there is limited information on the role of the microbiome in NPC, so we assessed variations in the nasopharynx microbiota of patients with NPC relative to the bacterial in health controls.
Methods
Nasopharynx lavage fluid (NLF) samples were collected from 11 NPC patients and 5 volunteer controls. 16S rRNA sequencing and comparative analyses of NLF bacterial microbiome between NPC patients and controls were performed.
Results
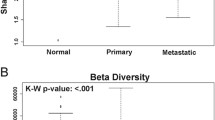
NLF microbial alpha-diversity by the Shannon index and Simpson index decreased significantly in the NPC patients when compared with the controls. Beta-diversity by principal component analysis exhibited separated patterns of the NPC patients and healthy controls. Thirty-one genera differed significantly between the NPC patient group and healthy control group. The abundance of 17 bacteria was correlated with primary tumor size and invaded lymph node size. Functional gene prediction analysis showed that 9 gene function pathways were significantly different between the two groups.
Conclusion
Our results demonstrated that the nasopharynx microbiota in NPC patients was different from that of the healthy controls, suggesting that the nasopharynx microenvironment might be related to NPC.



Similar content being viewed by others
Data availability
The data of this study are available by contacting the corresponding author upon reasonable request.
Code availability
Not applicable.
References
Siegel RL, Miller KD (2015) Jemal A (2015) Cancer statistics. CA Cancer J Clin 65(1):5–29. https://doi.org/10.3322/caac.21254
da Costa VG, Marques-Silva AC, Moreli ML (2015) The Epstein-Barr virus latent membrane protein-1 (LMP1) 30-bp deletion and XhoI-polymorphism in nasopharyngeal carcinoma: a meta-analysis of observational studies. Syst Rev 4:46. https://doi.org/10.1186/s13643-015-0037-z
Yoshizaki T, Ito M, Murono S, Wakisaka N, Kondo S, Endo K (2012) Current understanding and management of nasopharyngeal carcinoma. Auris Nasus Larynx 39(2):137–144. https://doi.org/10.1016/j.anl.2011.02.012
Garrett WS (2015) Cancer and the microbiota. Science 348(6230):80–86. https://doi.org/10.1126/science.aaa4972
Meurman JH (2010) Oral microbiota and cancer. J Oral Microbiol 2:5195. https://doi.org/10.3402/jom.v2i0.5195
Xu Y, Teng F, Huang S, Lin Z, Yuan X, Zeng X, Yang F (2014) Changes of saliva microbiota in nasopharyngeal carcinoma patients under chemoradiation therapy. Arch Oral Biol 59(2):176–186. https://doi.org/10.1016/j.archoralbio.2013.10.011
Zhu XX, Yang XJ, Chao YL, Zheng HM, Sheng HF, Liu HY, He Y, Zhou HW (2017) The Potential Effect of Oral Microbiota in the Prediction of Mucositis During Radiotherapy for Nasopharyngeal Carcinoma. EBioMedicine 18:23–31. https://doi.org/10.1016/j.ebiom.2017.02.002
Magoc T, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27(21):2957–2963. https://doi.org/10.1093/bioinformatics/btr507
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7(5):335–336. https://doi.org/10.1038/nmeth.f.303
Allen EK, Koeppel AF, Hendley JO, Turner SD, Winther B, Sale MM (2014) Characterization of the nasopharyngeal microbiota in health and during rhinovirus challenge. Microbiome 2:22. https://doi.org/10.1186/2049-2618-2-22
Perez-Losada M, Alamri L, Crandall KA, Freishtat RJ (2017) Nasopharyngeal microbiome diversity changes over time in children with asthma. PLoS ONE 12(1):e0170543. https://doi.org/10.1371/journal.pone.0170543
Mok SF, Karuthan C, Cheah YK, Ngeow WC, Rosnah Z, Yap SF, Ong HKA (2017) The oral microbiome community variations associated with normal, potentially malignant disorders and malignant lesions of the oral cavity. Malays J Pathol 39(1):1–15
Correa P (1992) Human gastric carcinogenesis: a multistep and multifactorial process–First American Cancer Society Award Lecture on Cancer Epidemiology and Prevention. Can Res 52(24):6735–6740
Nakatsu G, Li X, Zhou H, Sheng J, Wong SH, Wu WK, Ng SC, Tsoi H, Dong Y, Zhang N, He Y, Kang Q, Cao L, Wang K, Zhang J, Liang Q, Yu J, Sung JJ (2015) Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat Commun 6:8727. https://doi.org/10.1038/ncomms9727
Mitra A, MacIntyre DA, Marchesi JR, Lee YS, Bennett PR, Kyrgiou M (2016) The vaginal microbiota, human papillomavirus infection and cervical intraepithelial neoplasia: what do we know and where are we going next? Microbiome 4(1):58. https://doi.org/10.1186/s40168-016-0203-0
Liu HX, Tao LL, Zhang J, Zhu YG, Zheng Y, Liu D, Zhou M, Ke H, Shi MM, Qu JM (2018) Difference of lower airway microbiome in bilateral protected specimen brush between lung cancer patients with unilateral lobar masses and control subjects. Int J Cancer 142(4):769–778. https://doi.org/10.1002/ijc.31098
Said HS, Suda W, Nakagome S, Chinen H, Oshima K, Kim S, Kimura R, Iraha A, Ishida H, Fujita J, Mano S, Morita H, Dohi T, Oota H, Hattori M (2014) Dysbiosis of salivary microbiota in inflammatory bowel disease and its association with oral immunological biomarkers. DNA Res 21(1):15–25. https://doi.org/10.1093/dnares/dst037
Farrell JJ, Zhang L, Zhou H, Chia D, Elashoff D, Akin D, Paster BJ, Joshipura K, Wong DT (2012) Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut 61(4):582–588. https://doi.org/10.1136/gutjnl-2011-300784
Hartwig SM, Ketterer M, Apicella MA, Varga SM (2016) Non-typeable Haemophilus influenzae protects human airway epithelial cells from a subsequent respiratory syncytial virus challenge. Virology 498:128–135. https://doi.org/10.1016/j.virol.2016.08.020
Erwin AL, Smith AL (2007) Nontypeable Haemophilus influenzae: understanding virulence and commensal behavior. Trends Microbiol 15(8):355–362. https://doi.org/10.1016/j.tim.2007.06.004
Zaura E, Keijser BJ, Huse SM, Crielaard W (2009) Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol 9:259. https://doi.org/10.1186/1471-2180-9-259
Charlson ES, Chen J, Custers-Allen R, Bittinger K, Li H, Sinha R, Hwang J, Bushman FD, Collman RG (2010) Disordered microbial communities in the upper respiratory tract of cigarette smokers. PLoS ONE 5(12):e15216. https://doi.org/10.1371/journal.pone.0015216
Rocco CJ, Bakaletz LO, Goodman SD (2018) Targeting the HUbeta protein prevents Porphyromonas gingivalis from entering into pre-existing biofilms. J Bacteriol. https://doi.org/10.1128/JB.00790-17
Chavan RS, Pannaraj PS, Luna RA, Szabo S, Adesina A, Versalovic J, Krance RA, Kennedy-Nasser AA (2013) Significant morbidity and mortality attributable to rothia mucilaginosa infections in children with hematological malignancies or following hematopoietic stem cell transplantation. Pediatr Hematol Oncol 30(5):445–454. https://doi.org/10.3109/08880018.2013.783893
Eribe ERK, Olsen I (2017) Leptotrichia species in human infections II. J Oral Microbiol 9(1):1368848. https://doi.org/10.1080/20002297.2017.1368848
Vuotto F, Berthon C, Lemaitre N, Duhamel A, Balkaran S, Le Ray E, Micol JB, Faure K, Alfandari S (2013) Risk factors, clinical features, and outcome of Pseudomonas aeruginosa bacteremia in patients with hematologic malignancies: a case-control study. Am J Infect Control 41(6):527–530. https://doi.org/10.1016/j.ajic.2012.07.012
Stoodley BJ, Thom BT (1970) Observations on the intestinal carriage of Pseudomonas aeruginosa. J Med Microbiol 3(3):367–375. https://doi.org/10.1099/00222615-3-3-367
Funding
This study was supported by grants from the National Natural Science Foundation of China grant number No. 81603423 and No. 81601357, Dalian Science and Technology Star Project grant number 2017RQ040, and Liaoning Province Natural Science Foundation grant number 2019-ZD-0939.
Author information
Authors and Affiliations
Contributions
Yanjie GUO contributed to the study design. Longjie Li performed experiments and wrote the manuscript. Xiaoqin Deng and Yang Zou collected clinical sample. XiuPeng Lv and Longjie Li contributed to the data analyses and wrote the manuscript.
Corresponding author
Ethics declarations
Ethics approval
This study was approved by the ethics committee of the First Affiliated Hospital of Dalian Medical University.
Consent to participate
Written consent was obtained from all patients or the legal guardian of the patient to provide information and samples for research purposes.
Consent for publication
All authors read and approved the final manuscript.
Conflict of interest
The authors declare no competing interests.
Additional information
Responsible Editor: Jorge Luiz Mello Sampaio
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Li, L., Deng, X., Zou, Y. et al. Characterization of the nasopharynx microbiota in patients with nasopharyngeal carcinoma vs. healthy controls. Braz J Microbiol 52, 1873–1880 (2021). https://doi.org/10.1007/s42770-021-00594-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-021-00594-0