Abstract
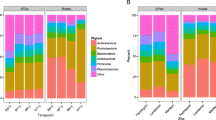
Poultry litter is widely applied as agricultural fertilizer and can affect the soil microbiome through nutrient overload and antibiotic contamination. In this study, we assessed changes in soil bacterial diversity using high-throughput sequencing approaches. Four samples in triplicate were studied: soils with short- and long-term fertilization by poultry litter (S1 = 10 months and S2 = 30 years, respectively), a soil inside a poultry shed (S3), and a forest soil used as control (S0). Samples S0, S1, and S2 revealed a relatively high richness, with confirmed operational taxonomic units (OTUs) in the three replicates of each sample ranging from 1243 to 1279, while richness in S3 was about three times lower (466). The most abundant phyla were Proteobacteria, Bacteroidetes, and Actinobacteria. Acidobacteria, Planctomycetes, and Verrucomicrobia were also abundant but highly diminished in S3, while Firmicutes was less abundant in S0. Changes in bacterial communities were very evident at the genera level. The genera Gaiella, Rhodoplanes, Solirubacter, and Sphingomonas were predominant in S0 but strongly decreased in the other soils. Pedobacter and Devosia were the most abundant in S1 and were diminished in S2, while Herbiconiux, Brevundimonas, Proteiniphilum, and Petrimonas were abundant in S2. The most abundant genera in S3 were Deinococcus, Truepera, Rhodanobacter, and Castellaniella. A predictive analysis of the metabolic functions with Tax4Fun2 software suggested the potential presence of enzymes associated with antibiotic resistance as well as with denitrification pathways, indicating that the S3 soil is a potential source of nitrous oxide, a powerful greenhouse gas.






Similar content being viewed by others
Data availability
The raw sequences were deposited in the Sequence Read Archive (SRA) database of National Center for Biotechnology Information (NCBI) - project PRJNA577278.
References
Fierer N (2017) Embracing the unknown: disentangling the complexities of the soil microbiome. Nat Rev Microbiol 15:579–590. https://doi.org/10.1038/nrmicro.2017.87
Jansson JK, Hofmockel KS (2018) The soil microbiome — from metagenomics to metaphenomics. Curr Opin Microbiol 43:162–168. https://doi.org/10.1016/j.mib.2018.01.013
Kaiser K, Wemheuer B, Korolkow V, Wemheuer F, Nacke H, Schöning I, Schrumpf M, Daniel R (2016) Driving forces of soil bacterial community structure, diversity, and function in temperate grasslands and forests. Sci Rep 6:1–12. https://doi.org/10.1038/srep33696
Rodríguez-Eugenio N, McLaughlin M, Pennock D (2018) Soil pollution: a hidden reality. Rome, FAO, p 142
Parente CET, Brito EMS, Azeredo A et al (2019) Fluoroquinolone antibiotics and their interactions in agricultural soils - a review. Orbital 11:42–52. https://doi.org/10.17807/orbital.v11i1.1352
Parente CET, Azeredo A, Vollú RE, Zonta E, Azevedo-Silva CE, Brito EMS, Seldin L, Torres JPM, Meire RO, Malm O (2019) Fluoroquinolones in agricultural soils: multi-temporal variation and risks in Rio de Janeiro upland region. Chemosphere 219:409–417. https://doi.org/10.1016/j.chemosphere.2018.11.184
Vollú RE, Cotta SR, Jurelevicius D, Leite DCA, Parente CET, Malm O, Martins DC, Resende ÁV, Marriel IE, Seldin L (2018) Response of the bacterial communities associated with maize rhizosphere to poultry litter as an organomineral fertilizer. Front Environ Sci 6:118. https://doi.org/10.3389/fenvs.2018.00118
Parente CET, Brusdzenski GS, Zonta E, Lino AS, Azevedo-Silva CE, Dorneles PR, Azeredo A, Torres JPM, Meire RO, Malm O (2020) Fluoroquinolones and trace elements in poultry litter: estimation of environmental load based on nitrogen requirement for crops. J Environ Sci Health B 0:1–12. https://doi.org/10.1080/03601234.2020.1816794
Freire LR, Balieiro FC, Zonta E et al (2013) Manual de calagem e adubação do Estado do Rio de Janeiro. Brasília, DF: Embrapa; Seropédica, RJ: Editora Universidade Rural. 430
Hashimoto GL, Abe Y, Sugita S (2007) The chemical composition of the early terrestrial atmosphere: formation of a reducing atmosphere from CI-like material. J Geophys Res E Planets 112:1–12. https://doi.org/10.1029/2006JE002844
Galloway JN, Dentener FJ, Capone DG, Boyer EW, Howarth RW, Seitzinger SP, Asner GP, Cleveland CC, Green PA, Holland EA, Karl DM, Michaels AF, Porter JH, Townsend AR, Vosmarty CJ (2004) Nitrogen cycles: past, present and future. Biogeochemistry 70:153–226. https://doi.org/10.1007/s10533-004-0370-0
Ren Z, Gao H, Elser JJ, Zhao Q (2017) Microbial functional genes elucidate environmental drivers of biofilm metabolism in glacier-fed streams. Sci Rep 7:1–8. https://doi.org/10.1038/s41598-017-13086-9
Parente CET, Lino AS, Arruda Junior ER, Zonta E, Dorneles PR, Torres JPM, Meire RO, Malm O (2019) Multi-temporal accumulation and risk assessment of available heavy metals in poultry litter fertilized soils from Rio de Janeiro upland region. Environ Monit Assess 191:28. https://doi.org/10.1007/s10661-018-7156-7
Bin HY, Zakaria MP, Latif PA, Saari N (2014) Occurrence of veterinary antibiotics and progesterone in broiler manure and agricultural soil in Malaysia. Sci Total Environ 488–489:261–267. https://doi.org/10.1016/j.scitotenv.2014.04.109
Ashworth AJ, DeBruyn JM, Allen FL et al (2017) Microbial community structure is affected by cropping sequences and poultry litter under long-term no-tillage. Soil Biol Biochem 114:210–219. https://doi.org/10.1016/j.soilbio.2017.07.019
Wei S, Morrison M, Yu Z (2013) Bacterial census of poultry intestinal microbiome. Poult Sci 92:671–683. https://doi.org/10.3382/ps.2012-02822
Feye KM, Baxter MFA, Tellez-Isaias G, Kogut MH, Ricke SC (2020) Influential factors on the composition of the conventionally raised broiler gastrointestinal microbiomes. Poult Sci 99:653–659. https://doi.org/10.1016/j.psj.2019.12.013
Liu P, Jia S, He X, Zhang X, Ye L (2017) Different impacts of manure and chemical fertilizers on bacterial community structure and antibiotic resistance genes in arable soils. Chemosphere 188:455–464. https://doi.org/10.1016/j.chemosphere.2017.08.162
Jangid K, Williams MA, Franzluebbers AJ, Sanderlin JS, Reeves JH, Jenkins MB, Endale DM, Coleman DC, Whitman WB (2008) Relative impacts of land-use, management intensity and fertilization upon soil microbial community structure in agricultural systems. Soil Biol Biochem 40:2843–2853. https://doi.org/10.1016/j.soilbio.2008.07.030
Santos HG, Jacomine PKT, Anjos LHC et al. (2018) Sistema brasileiro de classificação de solos. 5a edição, rev. e ampl. − Brasília, DF: Embrapa, 356
USDA – United States Department of Agriculture. Natural Resources Conservation Service. (1999) Soil taxonomy: a basic system of soil classification for making and interpreting soil surveys. 2.ed
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Press, Cold Spring Harbor Laboratory, New York
Andrews S (2016) FastQC: a quality control tool for high throughput sequence data. Babraham Bioinformatics. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed 18 Nov 2020
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. https://doi.org/10.1093/bioinformatics/btq461
Edgar RC (2016) UNOISE2: improved error-correction for Illumina 16S and ITS amplicon sequencing. bioRxiv 081257. https://doi.org/10.1101/081257
Shannon CE, Weaver W (1949) The mathematical theory of communication. The University of Illinois Press. 117
Simpson EH (1949) Measurement of diversity. Nature 163:688
Kim BR, Shin J, Guevarra RB, Lee JH, Kim DW, Seol KH, Lee JH, Kim HB, Isaacson RE (2017) Deciphering diversity indices for a better understanding of microbial communities. J Microbiol Biotechnol 27:2089–2093
Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31:814–821. https://doi.org/10.1038/nbt.2676
Aßhauer KP, Wemheuer B, Daniel R, Meinicke P (2015) Tax4Fun: predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics 31(17):2882–2884. https://doi.org/10.1093/bioinformatics/btv287
Wemheuer F, Taylor JA, Daniel R et al (2018) Tax4Fun2: a R-based tool for the rapid prediction of habitat-specific functional profiles and functional redundancy based on 16S rRNA gene marker gene sequences. bioRxiv 490037. https://doi.org/10.1101/490037
Koo H, Hakim JA, Morrow CD, Eipers PG, Davila A, Andersen DT, Bej AK (2017) Comparison of two bioinformatics tools used to characterize the microbial diversity and predictive functional attributes of microbial mats from Lake Obersee, Antarctica. J Microbiol Methods 140:15–22. https://doi.org/10.1016/j.mimet.2017.06.017
Zhu Y, Cao Y, Yang M, Wen P, Cao L, Ma J, Zhang Z, Zhang W (2018) Bacterial diversity and community in Qula from the Qinghai–Tibetan Plateau in China. PeerJ 2018:1–22. https://doi.org/10.7717/peerj.6044
Shange RS, Ankumah RO, Zabawa R, Dowd SE (2013) Bacterial community structure and composition in soils under industrial poultry production activities: an observational study. Air Soil Water Res 6:91–101. https://doi.org/10.4137/ASWR.S12009
Zhen Z, Liu H, Wang N, Guo L, Meng J, Ding N, Wu G, Jiang G (2014) Effects of manure compost application on soil microbial community diversity and soil microenvironments in a temperate cropland in China. PLoS One 9(10):e108555. https://doi.org/10.1371/journal.pone.0108555
Choi S, Kim S, Shin JY, Kim MK, Kim JH (2015) Development and verification for analysis of pesticides in eggs and egg products using QuEChERS and LC-MS/MS. Food Chem 173:1236–1242. https://doi.org/10.1016/j.foodchem.2014.10.143
Sood U, Gupta V, Kumar R, Lal S, Fawcett D, Rattan S, Poinern GEJ, Lal R (2020) Chicken gut microbiome and human health: past scenarios, current perspectives, and futuristic applications. Indian J Microbiol 60:2–11. https://doi.org/10.1007/s12088-019-00785-2
Zou A, Sharif S, Parkinson J (2018) Lactobacillus elicits a ‘Marmite effect’ on the chicken cecal microbiome. npj Biofilms Microbiomes 4:1–5. https://doi.org/10.1038/s41522-018-0070-5
Theodorakopoulos N, Bachar D, Christen R, Alain K, Chapon V (2013) Exploration of Deinococcus-Thermus molecular diversity by novel group-specific PCR primers. Microbiologyopen 2:862–872. https://doi.org/10.1002/mbo3.119
Wei H, Wang L, Hassan M, Xie B (2018) Succession of the functional microbial communities and the metabolic functions in maize straw composting process. Bioresour Technol 256:333–341. https://doi.org/10.1016/j.biortech.2018.02.050
de Gannes V, Eudoxie G, Hickey WJ (2013) Prokaryotic successions and diversity in composts as revealed by 454-pyrosequencing. Bioresour Technol 133:573–580. https://doi.org/10.1016/j.biortech.2013.01.138
Chen P, Zhang C, Ju X, Xiong Y, Xing K, Qin S (2019) Community composition and metabolic potential of endophytic actinobacteria from coastal salt marsh plants in Jiangsu, China. Front Microbiol 10:1–16. https://doi.org/10.3389/fmicb.2019.01063
Pérez ML, Collavino MM, Sansberro PA, Mroginski LA, Galdeano E (2016) Diversity of endophytic fungal and bacterial communities in Ilex paraguariensis grown under field conditions. World J Microbiol Biotechnol 32:1–15. https://doi.org/10.1007/s11274-016-2016-5
Ryan MP, Pembroke JT (2018) Brevundimonas spp: emerging global opportunistic pathogens. Virulence 9:480–493. https://doi.org/10.1080/21505594.2017.1419116
Velho-Pereira S, Kamat NM (2011) Antimicrobial screening of actinobacteria using a modified cross-streak method. Indian J Pharm Sci 73:223–228. https://doi.org/10.4103/0250-474x.91566
Lazzarini A, Cavaletti L, Toppo G, Marinelli F (2001) Rare genera of actinomycetes as potential producers of new antibiotics. A Van Leeuw 79:219–405. https://doi.org/10.1023/A:1010287600557
Anhalt JC, Moorman TB, Koskinen WC (2007) Biodegradation of imidacloprid by an isolated soil microorganism. J Environ Sci Heal Part B 42:509–514. https://doi.org/10.1080/03601230701391401
Chen Q, An X, Li H, Su J, Ma Y, Zhu YG (2016) Long-term field application of sewage sludge increases the abundance of antibiotic resistance genes in soil. Environ Int 92–93:1–10. https://doi.org/10.1016/j.envint.2016.03.026
Rogeri DA, Ernani PR, Mantovani A, Lourenço KS (2016) Composition of poultry litter in southern Brazil. Rev Bras Cienc do Solo 40:1–7. https://doi.org/10.1590/18069657rbcs20140697
Sait M, Davis KER, Janssen PH (2006) Effect of pH on isolation and distribution of members of subdivision 1 of the phylum acidobacteria occurring in soil. Appl Env Microbiol 72:1852–1857. https://doi.org/10.1128/AEM.72.3.1852
Gupta RS (2000) The phylogeny of proteobacteria: relationships to other eubacterial phyla and eukaryotes. FEMS Microbiol Rev 24:367–402. https://doi.org/10.1016/S0168-6445(00)00031-0
Sistani KR, Jn-Baptiste M, Lovanh N, Cook KL (2011) atmospheric emissions of nitrous oxide, methane, and carbon dioxide from different nitrogen fertilizers. J Environ Qual 40:1797–1805. https://doi.org/10.2134/jeq2011.0197
Posmanik R, Nejidat A, Dahan O, Gross A (2017) Seasonal and soil-type dependent emissions of nitrous oxide from irrigated desert soils amended with digested poultry manures. Sci Total Environ 593–594:91–98. https://doi.org/10.1016/j.scitotenv.2017.03.115
Davis BW, Mirsky SB, Needelman BA, Cavigelli MA, Yarwood SA (2019) Nitrous oxide emissions increase exponentially with organic N rate from cover crops and applied poultry litter. Agric Ecosyst Environ 272:165–174. https://doi.org/10.1016/j.agee.2018.10.023
Braker G, Fesefeldt A, Witzel KP (1998) Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl Environ Microbiol 64:3769–3775. https://doi.org/10.1128/aem.64.10.3769-3775.1998
Philippot L, Piutti S, Martin-Laurent F et al (2002) Molecular analysis of the nitrate-reducing community from unplanted and maize-planted soils. Appl Environ Microbiol 68:6121–6128. https://doi.org/10.1128/AEM.68.12.6121-6128.2002
Heuer H, Schmitt H, Smalla K (2011) Antibiotic resistance gene spread due to manure application on agricultural fields. Curr Opin Microbiol 14:236–243. https://doi.org/10.1016/j.mib.2011.04.009
Cytryn E (2013) The soil resistome: The anthropogenic, the native, and the unknown. Soil Biol Biochem 63:18–23. https://doi.org/10.1016/j.soilbio.2013.03.017
Van Goethem MW, Pierneef R, Bezuidt OKI et al (2018) A reservoir of ‘historical’ antibiotic resistance genes in remote pristine Antarctic soils. Microbiome 6:1–12. https://doi.org/10.1186/s40168-018-0424-5
Li C, Chen J, Wang J, Ma Z, Han P, Luan Y, Lu A (2015) Occurrence of antibiotics in soils and manures from greenhouse vegetable production bases of Beijing, China and an associated risk assessment. Sci Total Environ 521–522:101–107. https://doi.org/10.1016/j.scitotenv.2015.03.070
Sun J, Zeng Q, Tsang DCW, Zhu LZ, Li XD (2017) Antibiotics in the agricultural soils from the Yangtze River Delta, China. Chemosphere 189:301–308. https://doi.org/10.1016/j.chemosphere.2017.09.040
Perry JA, Westman EL, Wright GD (2014) The antibiotic resistome: what’s new? Curr Opin Microbiol 21:45–50. https://doi.org/10.1016/j.mib.2014.09.002
Parente CET, Sierra J, Martí E (2018) Ecotoxicity and biodegradability of oxytetracycline and ciprofloxacin on terrestrial and aquatic media. Orbital 10:262–271. https://doi.org/10.17807/orbital.v10i4.1063
Roberts CM, Schwarz S (2009) Tetracycline and chloramphenicol resistance mechanisms. In: Mayers D.L. (eds) Antimicrobial drug resistance. Infectious Disease. Humana Press. pp 183–193. https://doi.org/10.1007/978-1-59745-180-2_15
Nesme J, Cécillon S, Delmont TO et al (2014) Large-scale metagenomic-based study of antibiotic resistance in the environment. Curr Biol 24:1096–1100. https://doi.org/10.1016/j.cub.2014.03.036
Zapun A, Contreras-Martel C, Vernet T (2008) Penicillin-binding proteins and β-lactam resistance. FEMS Microbiol Rev 32:361–385. https://doi.org/10.1111/j.1574-6976.2007.00095.x
Udikovic-Kolic N, Wichmann F, Broderick NA, Handelsman J (2014) Bloom of resident antibiotic-resistant bacteria in soil following manure fertilization. Proc Natl Acad Sci USA 111:15202–15207. https://doi.org/10.1073/pnas.1409836111
Podnecky NL, Wuthiekanun V, Peacock SJ, Schweizer HP (2013) The BpeEF-OprC efflux pump is responsible for widespread trimethoprim resistance in clinical and environmental Burkholderia pseudomallei isolates. Antimicrob Agents Chemother 57:4381–4386. https://doi.org/10.1128/AAC.00660-13
Rusu A, Hancu G, Uivaroşi V (2014) Fluoroquinolone pollution of food, water and soil, and bacterial resistance. Environ Chem Lett 13:21–36. https://doi.org/10.1007/s10311-014-0481-3
Blanco P, Hernando-Amado S, Reales-Calderon J, Corona F, Lira F, Alcalde-Rico M, Bernardini A, Sanchez M, Martinez J (2016) Bacterial multidrug efflux pumps: much more than antibiotic resistance determinants. Microorganisms 4:14. https://doi.org/10.3390/microorganisms4010014
Acknowledgements
The authors would like to thank the poultry farmers from SJVRP for their essential contribution to the work.
Code availability
Accession numbers SAMN13023011–SAMN13023014.
Funding
This work was supported by the Brazilian Ministry of Science, Technology, Innovations and Communications/National Council for Scientific and Technological Development (MCTI/CNPq-Universal-01/2016), process 426192/2016-8 and Dirección de Apoyo a la Investigación y al Posgrado (DAIP, Universidad de Guanajuato-UG, project 206/2019, in Mexico. C.E.T.P. has a PNPD grant from Brazilian Foundation for the Coordination and Improvement of Higher Level of Education Personnel (CAPES); E.A.C.R and A.P.F.C. received undergraduate scholarships from DAIP-UG; O.M. has a grant “Cientista do Nosso Estado” from Carlos Chagas Filho Research Support Foundation of Rio de Janeiro State (FAPERJ).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by Cláudio Ernesto Taveira Parente, Elcia Margareth Souza Brito, César Augusto Caretta, Renata Estebanez Vollú, and Lucy Seldin. The first draft of the manuscript was written by Cláudio Ernesto Taveira Parente, Elcia Margareth Souza Brito, and César Augusto Caretta. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Responsible Editor: Melissa Fontes Landell.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 2032 kb)
Rights and permissions
About this article
Cite this article
Parente, C.E.T., Brito, E.M.S., Caretta, C.A. et al. Bacterial diversity changes in agricultural soils influenced by poultry litter fertilization. Braz J Microbiol 52, 675–686 (2021). https://doi.org/10.1007/s42770-021-00437-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-021-00437-y