Abstract
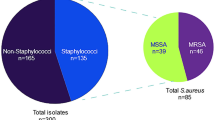
This study aimed to evaluate virulence factors and genetic markers of antimicrobial resistance in 400 Staphylococcus aureus strains isolated from bovine mastitis in four Brazilian states, as well as to assess the association between these characteristics and field information. Virulence factors and drug resistance genes were identified by PCR screening. Biofilm-forming and hemolytic phenotype were detected using Congo red Tryptic Soy Broth and defibrinated sheep blood agar, respectively. Of all isolates, 83.5% were biofilm-forming and 98.5% strains exhibited biofilm gene icaAD, and a significant association between phenotype and genotype for biofilm was observed (P = 0.0005). Hemolysin genes were observed in 82.85% (hla+hlb+), 16.5% (hla+) and 0.75% (hlb+) isolates, whereas the hemolytic phenotype exhibited was complete and incomplete hemolysis in 64.25%, complete in 28.25%, incomplete in 4.75%, and negative in 2.75% of the strains. Virulence factors genes luk, seb, sec, sed, and tst were observed in 3.5%, 0.5%, 1%, 0.25%, and 0.74% isolates, respectively. The gene blaZ was detected in 82.03% of penicillin-resistant isolates, whereas tetK and aac(6′)-Ie–aph(2′)-Ia were observed in 33.87% and 45.15% of the tetracycline and aminoglycosides-resistant isolates, respectively. Fluoroquinolone resistance gene mepA was detected for the first time in S. aureus from bovine mastitis. Resistance genes tetM (3.22%), tetL (1.61%), ermA (14.29%), ermB (14.29%), ermC (33.3%), ermT (9.52%), ermY (4.76%), msrA (9.52%), and mphC (9.52%) were also detected among resistant isolates. No association between virulence factors or antimicrobial-resistant genes and year of isolation, geographic origin, or antimicrobial resistance profile was observed. Our results showed that S. aureus strains isolated from bovine mastitis in the four Brazilian states sampled are mainly biofilm-forming and hemolytic, whereas virulence genes associated with enterotoxins, luk and tst, were less frequently observed. Moreover, a wide variety of resistance genes that confer resistance to almost all classes of antimicrobial agents approved for use in animals and humans were found. Overall, the data point to a great pathogenic potential of S. aureus associated with bovine mastitis and to the non-negligible risks to public health of staphylococcal infections from animal origin.


Similar content being viewed by others
References
Ruegg PL (2017) A 100-year review: mastitis detection, management, and prevention. J Dairy Sci 100(12):10381–10397. https://doi.org/10.3168/jds.2017-13023
Cortimiglia C, Luini M, Bianchini V, Marzagalli L, Vezzoli F, Avisani D, Bertoletti M, Ianzano A, Franco A, Battisti A (2016) Prevalence of Staphylococcus aureus and of methicillin-resistant S. aureus clonal complexes in bulk tank milk from dairy cattle herds in Lombardy Region (Northern Italy). Epidemiol Infect 144(14):3046–3051. https://doi.org/10.1017/S0950268816001576
Abebe R, Hatiya H, Abera M, Megersa B, Asmare K (2016) Bovine mastitis: prevalence, risk factors and isolation of Staphylococcus aureus in dairy herds at Hawassa milk shed, South Ethiopia. BMC Vet Res 12(1):270. https://doi.org/10.1186/s12917-016-0905-3
Olde Riekerink RG, Barkema HW, Scholl DT, Poole DE, Kelton DF (2010) Management practices associated with the bulk-milk prevalence of Staphylococcus aureus in Canadian dairy farms. Prev Vet Med 97(1):20–28. https://doi.org/10.1016/j.prevetmed.2010.07.002
Marques VF, Motta CC, Soares BD, Melo DA, Coelho SM, Coelho ID, Barbosa HS, Souza MM (2017) Biofilm production and beta-lactamic resistance in Brazilian Staphylococcus aureus isolates from bovine mastitis. Braz J Microbiol 48(1):118–124. https://doi.org/10.1016/j.bjm.2016.10.001
Silveira-Filho VM, Luz IS, Campos AP, Silva WM, Barros MP, Medeiros ES, Freitas MF, Mota RA, Sena MJ, Leal-Balbino TC (2014) Antibiotic resistance and molecular analysis of Staphylococcus aureus isolated from cow's milk and dairy products in northeast Brazil. J Food Protect 77(4):583–591. https://doi.org/10.4315/0362-028x.jfp-13-343
Araújo RMP, Peixoto RM, Peixoto LJS, Gouveia GV, Costa MM (2017) Virulence factors in Staphylococcus aureus and quality of raw milk from dairy cows in a semiarid region of northeastern Brazil. Acta Sci Vet 45(1491):10.22456/1679–10.9216.80637
Rall V, Miranda E, Castilho I, Camargo C, Langoni H, Guimarães F, Júnior JA, Júnior AF (2014) Diversity of Staphylococcus species and prevalence of enterotoxin genes isolated from milk of healthy cows and cows with subclinical mastitis. J Dairy Sci 97(2):829–837. https://doi.org/10.3168/jds.2013-7226
Lina G, Quaglia A, Reverdy ME, Leclercq R, Vandenesch F, Etienne J (1999) Distribution of genes encoding resistance to macrolides, lincosamides, and streptogramins among staphylococci. Antimicrob Agents Chemother 43(5):1062–1066
Jarraud S, Mougel C, Thioulouse J, Lina G, Meugnier H, Forey F, Nesme X, Etienne J, Vandenesch F (2002) Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect Immun 70(2):631–641. https://doi.org/10.1128/IAI.70.2.631-641.2002
Sun F, Wang Q, Xia P (2003) PCR analysis of clinically isolated Staphylococcus aureus biofilm associated gene. Acta Acad Med Militaris Tertiae 31(15):1147–1149
Boynukara B, Gulhan T, Alisarli M, Gurturk K, Solmaz H (2008) Classical enterotoxigenic characteristics of Staphylococcus aureus strains isolated from bovine subclinical mastitis in Van, Turkey. Int J Food Microbiol 125(2):209–211. https://doi.org/10.1016/j.ijfoodmicro.2008.03.024
Shrivastava N, Sharma V, Shrivastav A, Nayak A, Rai AK (2018) Prevalence and characterization of Panton-Valentine leukocidin-positive Staphylococcus aureus in bovine milk in Jabalpur district of Madhya Pradesh, India. Vet World 11(3):316. https://doi.org/10.14202/vetworld.2018.316-320
Tam K, Torres VJ (2019) Staphylococcus aureus secreted toxins and extracellular enzymes. Microbiol Spectrum 7(2). https://doi.org/10.1128/microbiolspec.GPP3-0039-2018
Cucarella C, Tormo MÁ, Úbeda C, Trotonda MP, Monzón M, Peris C, Amorena B, Lasa Í, Penadés JR (2004) Role of biofilm-associated protein bap in the pathogenesis of bovine Staphylococcus aureus. Infect Immun 72(4):2177–2185. https://doi.org/10.1128/IAI.72.4.2177-2185.2004
Kinross P, Petersen A, Skov R, Van Hauwermeiren E, Pantosti A, Laurent F, Voss A, Kluytmans J, Struelens MJ, Heuer O, Monnet DL (2017) Livestock-associated meticillin-resistant Staphylococcus aureus (MRSA) among human MRSA isolates, European Union/European economic area countries, 2013. Euro Surveill 22(44). https://doi.org/10.2807/1560-7917.es.2017.22.44.16-00696
Qu Y, Zhao H, Nobrega DB, Cobo ER, Han B, Zhao Z, Li S, Li M, Barkema HW, Gao J (2019) Molecular epidemiology and distribution of antimicrobial resistance genes of Staphylococcus species isolated from Chinese dairy cows with clinical mastitis. J Dairy Sci 102(2):1571–1583. https://doi.org/10.3168/jds.2018-15136
García-Álvarez L, Holden MT, Lindsay H, Webb CR, Brown DF, Curran MD, Walpole E, Brooks K, Pickard DJ, Teale C (2011) Methicillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect Dis 11(8):595–603. https://doi.org/10.1016/S1473-3099(11)70126-8
Becker K, Larsen AR, Skov RL, Paterson GK, Holmes MA, Sabat AJ, Friedrich AW, Köck R, Peters G, Kriegeskorte A (2013) Evaluation of a modular multiplex-PCR methicillin-resistant Staphylococcus aureus detection assay adapted for mecC detection. J Clin Microbiol 51(6):1917–1919. https://doi.org/10.1128/JCM.00075-13
Martini CL, Lange CC, Brito MA, Ribeiro JB, Mendonca LC, Vaz EK (2017) Characterisation of penicillin and tetracycline resistance in Staphylococcus aureus isolated from bovine milk samples in Minas Gerais, Brazil. J Dairy Res 84(2):202–205. https://doi.org/10.1017/S0022029917000061
Lüthje P, Schwarz S (2006) Antimicrobial resistance of coagulase-negative staphylococci from bovine subclinical mastitis with particular reference to macrolide–lincosamide resistance phenotypes and genotypes. J Antimicrob Chemother 57(5):966–969. https://doi.org/10.1093/jac/dkl061
Silva NC, Guimarães FF, Manzi MP, Júnior AF, Gómez-Sanz E, Gómez P, Langoni H, Rall VL, Torres C (2014) Methicillin-resistant Staphylococcus aureus of lineage ST398 as cause of mastitis in cows. Lett Appl Microbiol 59(6):665–669. https://doi.org/10.1111/lam.12329
Cretenet M, Even S, Le Loir Y (2011) Unveiling Staphylococcus aureus enterotoxin production in dairy products: a review of recent advances to face new challenges. J Dairy SciTechnol 91(2):127–150. https://doi.org/10.1007/s13594-011-0014-9
Markey B, Leonard F, Archambault M, Cullinane A, Maguire D (2013) Clinical veterinary microbiology E-book. Elsevier Health Sciences
Aizawa J, Souza-Filho AF, Guimarães AS, Vasconcelos CG, Brito MAV, Sellera FP, Cortez A, Heinemann MB (2019) Retrospective multicenter study reveals absence of MRSA-associated bovine mastitis in Brazil (1994 to 2016). J Infect Dev Countr 13(06):581–583. https://doi.org/10.3855/jidc.11406
Pitcher DG, Saunders NA, Owen RJ (1989) Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol 8(4):151–156. https://doi.org/10.1111/j.1472-765X.1989.tb00262.x
Cremonesi P, Luzzana M, Brasca M, Morandi S, Lodi R, Vimercati C, Agnellini D, Caramenti G, Moroni P, Castiglioni B (2005) Development of a multiplex PCR assay for the identification of Staphylococcus aureus enterotoxigenic strains isolated from milk and dairy products. Mol Cell Probes 19(5):299–305. https://doi.org/10.1016/j.mcp.2005.03.002
Mehrotra M, Wang G, Johnson WM (2000) Multiplex PCR for detection of genes for Staphylococcus aureus enterotoxins, exfoliative toxins, toxic shock syndrome toxin 1, and methicillin resistance. J Clin Microbiol 38(3):1032–1035. https://doi.org/10.1128/JCM.38.3.1032-1035.2000
Lina G, Piémont Y, Godail-Gamot F, Bes M, Peter M-O, Gauduchon V, Vandenesch F, Etienne J (1999) Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis 29(5):1128–1132. https://doi.org/10.1086/313461
Lee J-S, Bae Y-M, Han A, Lee S-Y (2016) Development of Congo red broth method for the detection of biofilm-forming or slime-producing Staphylococcus sp. LWT Food Sci Technol 73:707–714. https://doi.org/10.1016/j.lwt.2016.03.023
Schnellmann C, Gerber V, Rossano A, Jaquier V, Panchaud Y, Doherr MG, Thomann A, Straub R, Perreten V (2006) Presence of new mecA and mph(C) variants conferring antibiotic resistance in Staphylococcus spp. isolated from the skin of horses before and after clinic admission. J Clin Microbiol 44(12):4444–4454. https://doi.org/10.1128/jcm.00868-06
Sutcliffe J, Grebe T, Tait-Kamradt A, Wondrack L (1996) Detection of erythromycin-resistant determinants by PCR. Antimicrob Agents Chemother 40(11):2562–2566
Gomez-Sanz E, Torres C, Lozano C, Fernandez-Perez R, Aspiroz C, Ruiz-Larrea F, Zarazaga M (2010) Detection, molecular characterization, and clonal diversity of methicillin-resistant Staphylococcus aureus CC398 and CC97 in Spanish slaughter pigs of different age groups. Foodborne Pathog Dis 7(10):1269–1277. https://doi.org/10.1089/fpd.2010.0610
Aarestrup FM, Agersø Y, Ahrens P, Jørgensen JCØ, Madsen M, Jensen LB (2000) Antimicrobial susceptibility and presence of resistance genes in staphylococci from poultry. Vet Microbiol 74(4):353–364. https://doi.org/10.1016/S0378-1135(00)00197-8
Vakulenko SB, Donabedian SM, Voskresenskiy AM, Zervos MJ, Lerner SA, Chow JW (2003) Multiplex PCR for detection of aminoglycoside resistance genes in enterococci. Antimicrob Agents Chemother 47(4):1423–1426
Couto I, Costa SS, Viveiros M, Martins M, Amaral L (2008) Efflux-mediated response of Staphylococcus aureus exposed to ethidium bromide. J Antimicrob Chemother 62(3):504–513. https://doi.org/10.1093/jac/dkn217
Pan X-S, Hamlyn PJ, Talens-Visconti R, Alovero FL, Manzo RH, Fisher LM (2002) Small-colony mutants of Staphylococcus aureus allow selection of gyrase-mediated resistance to dual-target fluoroquinolones. Antimicrob Agents Chemother 46(8):2498–2506. https://doi.org/10.1128/AAC.46.8.2498-2506.2002
CLSI (2017) CLSI supplement M100, 27th edn. Clinical and Laboratory Standards Institute, Wayne
Ewing B, Hillier L, Wendl MC, Green P (1998) Base-calling of automated sequencer traces using phred. I Accuracy assessment. Genome Res 8(3):175–185
Huang X, Madan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9(9):868–877. https://doi.org/10.1101/gr.9.9.868
Yamagishi J, Kojima T, Oyamada Y, Fujimoto K, Hattori H, Nakamura S, Inoue M (1996) Alterations in the DNA topoisomerase IV grlA gene responsible for quinolone resistance in Staphylococcus aureus. Antimicrob Agents Chemother 40(5):1157–1163. https://doi.org/10.1128/AAC.40.5.1157
Ito H, Yoshida H, Bogaki-Shonai M, Niga T, Hattori H, Nakamura S (1994) Quinolone resistance mutations in the DNA gyrase gyrA and gyrB genes of Staphylococcus aureus. Antimicrob Agents Chemother 38(9):2014–2023. https://doi.org/10.1128/AAC.38.9.2014
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Castelani L, Pilon LE, Martins T, Pozzi CR, Arcaro JRP (2015) Investigation of biofilm production and ica A and ica D genes in Staphylococcus aureus isolated from heifers and cows with mastitis. J Anim Sci 86(3):340–344. https://doi.org/10.1111/asj.12284
Li L, Yang H, Liu D, He H, Wang C, Zhong J, Gao T, Zeng Y (2012) Analysis of biofilm formation and associated gene detection in Staphylococcus isolates from bovine mastitis. Afr J Biotechnol 11(8):2113–2118. https://doi.org/10.5897/AJB11.081
Vasudevan P, Nair MKM, Annamalai T, Venkitanarayanan KS (2003) Phenotypic and genotypic characterization of bovine mastitis isolates of Staphylococcus aureus for biofilm formation. Vet Microbiol 92(1):179–185. https://doi.org/10.1016/S0378-1135(02)00360-7
Tang JN, Zhou R, Wang HN, Zeng ZG (2008) Accessory gene regulator in Staphylococcus biofilm formation and infection. J Cent South Univ 33(11):1066–1070
Figueiredo AMS, Ferreira FA, Beltrame CO, Cortes MF (2017) The role of biofilms in persistent infections and factors involved in ica-independent biofilm development and gene regulation in Staphylococcus aureus. Crit Rev Microbiol 43(5):602–620. https://doi.org/10.1080/1040841X.2017.1282941
Yang FL, Li XS, Liang XW, Zhang XF, Qin GS, Yang BZ (2012) Detection of virulence-associated genes in Staphylococcus aureus isolated from bovine clinical mastitis milk samples in Guangxi. Trop Anim Health Prod 44(8):1821–1826. https://doi.org/10.1007/s11250-012-0143-z
Silva ER, Boechat JUD, Martins JCD, Ferreira WPB, Siqueira AP, da Silva N (2005) Hemolysin production by Staphylococcus aureus species isolated from mastitic goat milk in Brazilian dairy herds. Small Ruminant Res 56(1–3):271–275. https://doi.org/10.1016/j.smallrumres.2004.04.011
Pérez V, Costa G, Guimarães A, Heinemann M, Lage A, Dorneles E (2020) Relationship between virulence factors and antimicrobial resistance in Staphylococcus aureus from bovine mastitis. J Glob Antimicrob Res 22:792–802. https://doi.org/10.1016/j.jgar.2020.06.010
Cifrian E, Guidry A, Bramley A, Norcross N, Bastida-Corcuera F, Marquardt W (1996) Effect of staphylococcal β toxin on the cytotoxicity, proliferation and adherence of Staphylococcus aureus to bovine mammary epithelial cells. Vet Microbiol 48(3–4):187–198. https://doi.org/10.1016/0378-1135(95)00159-X
Hennekinne J-A, Ostyn A, Guillier F, Herbin S, Prufer A-L, Dragacci S (2010) How should staphylococcal food poisoning outbreaks be characterized? Toxins 2(8):2106–2116. https://doi.org/10.3390/toxins2082106
Liu Y, Chen W, Ali T, Alkasir R, Yin J, Liu G, Han B (2014) Staphylococcal enterotoxin H induced apoptosis of bovine mammary epithelial cells in vitro. Toxins 6(12):3552–3567. https://doi.org/10.3390/toxins6123552
Ruaro A, Andrighetto C, Torriani S, Lombardi A (2013) Biodiversity and characterization of indigenous coagulase-negative staphylococci isolated from raw milk and cheese of North Italy. Food Microbiol 34(1):106–111. https://doi.org/10.1016/j.fm.2012.11.013
Fitzgerald J, Hartigan P, Meaney W, Smyth C (2000) Molecular population and virulence factor analysis of Staphylococcus aureus from bovine intramammary infection. J Applied Microbiol 88(6):1028–1037. https://doi.org/10.1046/j.1365-2672.2000.01071.x
Fluit A (2012) Livestock-associated Staphylococcus aureus. Clin Microbiol Infect 18(8):735–744. https://doi.org/10.1111/j.1469-0691.2012.03846.x
Poole K (2007) Efflux pumps as antimicrobial resistance mechanisms. Ann Med 39(3):162–176. https://doi.org/10.1080/07853890701195262
Correia S, Poeta P, Hébraud M, Capelo JL, Igrejas G (2017) Mechanisms of quinolone action and resistance: Where do we stand? J Med Microbiol 66(5):551–559. https://doi.org/10.1099/jmm.0.000475
Silva JR, Castro GAC, Gonçalves MS, Custódio DAC, Mian GF, Costa GM (2017) In vitro antimicrobial susceptibility and genetic resistance determinants of Streptococcus agalactiae isolated from mastitic cows in Brazilian dairy herds. Semina: Ciênc Agrár 38(4):2581–2594. https://doi.org/10.5433/1679-0359.2017v38n4Supl1p2581
Acknowledgments
VCP is grateful to Capes and OAS (Organization of American States) for her fellowship. MBH and APL are thankful to CNPq for their fellowships.
Availability of data and material
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials.
Code availability
Not applicable
Funding
This study was supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação de Amparo à Pesquisa de Minas Gerais (Fapemig), Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP 2015/10332-6), and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest/competing interests
The authors declare that they have no conflict of interest.
Ethics approval
Not applicable.
Consent to participate
Not applicable
Consent for publication
All authors gave the consent for publication.
Additional information
Responsible Editor: Luis Augusto Nero.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Pérez, V.K.C., Custódio, D.A.C., Silva, E.M.M. et al. Virulence factors and antimicrobial resistance in Staphylococcus aureus isolated from bovine mastitis in Brazil. Braz J Microbiol 51, 2111–2122 (2020). https://doi.org/10.1007/s42770-020-00363-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-020-00363-5