Abstract
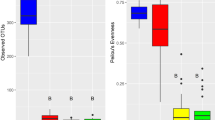
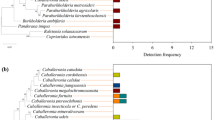
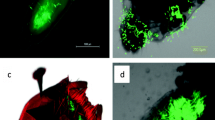
The development of insects is strongly influenced by their resident microorganisms. Symbionts play key roles in insect nutrition, reproduction, and defense. Bacteria are important partners due to the wide diversity of their biochemical pathways that aid in the host development. We present evidence that the foam produced by nymphs of the spittlebug Mahanarva fimbriolata harbors a diversity of bacteria, including some that were previously reported as defensive symbionts of insects. Analysis of the microbiomes in the nymph gut and the soil close to the foam showed that the microorganisms in the foam were more closely related to those in the gut than in the soil, suggesting that the bacteria are actively introduced into the foam by the insect. Proteobacteria, Actinobacteria, and Acidobacteria were the predominant groups found in the foam. Since members of Actinobacteria have been found to protect different species of insects by producing secondary metabolites with antibiotic properties, we speculate that the froth produced by M. fimbriolata may aid in defending the nymphs against entomopathogenic microorganisms.



Similar content being viewed by others
References
Guilbeau BH (1908) The origin and formation of the froth in spittle-insects. Am Nat 42:783–798
Kato K (1958) The origin and composition of the cuckoo spit. Rept Saitama Univ, B 3:33–53 https://ci.nii.ac.jp/naid/10026887170#cit.
Weaver C, King D (1954) Meadow spittlebug. Ohio Agric Exp Station Res Bull 741:1–99. https://scholar.google.com/scholar_lookup?title=Meadow+spittlebug,+Philaenus+leucophthalmus+(L.)&author=Weaver,+C.R.&author=King,+D.R.&publication_year=1954&journal=Ohio+Agric.+Exp.+Stat.+Bull.&volume=741&pages=1%E2%80%9399
Marshall AT (1973) Protein synthesis and secretion by the Malpighian tubules of cercopoid larvae (Homoptera). J Insect Physiol 19:2317–2326
Mello MLS, Pimentel ER, Yamada AT, Storopoli-Neto A (1987) Composition and structure of the froth of the spittlebug, Deois sp. Insect Biochem 17:493–502
Tonelli M, Gomes G, Silva WD, Magri NTC, Vieira DM, Aguiar CL et al (2018) Spittlebugs produce foam as a thermoregulatory adaptation. Sci Rep 8:4729. https://doi.org/10.1038/s41598-018-23031-z
Whittaker JB (1970) Cercopid spittle as a microhabitat. Oikos. 21:59–64. https://doi.org/10.2307/3543839
del Campo ML, King JT, Gronquist MR (2011) Defensive and chemical characterization of the froth produced by the cercopid Aphrophora cribrata. Chemoecology. 21:1–8. https://doi.org/10.1007/s00049-010-0059-x
Leite LG, Machado LA, Goulart RM, Tavares FM, Batista FA (2005) Screening of entomopathogenic nematodes (Nemata: Rhabditida) and the efficiency of Heterorhabditis sp. against the sugarcane root spittlebug Mahanarva fimbriolata (Fabr.) (Hemiptera: Cercopidae). Neotrop Entomol 34:785–790. https://doi.org/10.1590/S1519-566X2005000500010
Dinardo-Miranda LL, Vasconcelos ACM, Vieira SR, Fracasso JV, Grego CR (2007) Uso da geoestatística na avaliação da distribuição espacial de Mahanarva fimbriolata em cana-de-açúcar. Bragantia. 66:449–455
Rezende JM, Zanardo ABR, da Silva LM, Delalibera I, Rehner SA (2015) Phylogenetic diversity of Brazilian Metarhizium associated with sugarcane agriculture. BioControl. 60:495–505. https://doi.org/10.1007/s10526-015-9656-5
Ferrari J, Darby AC, Daniell TJ, Godfray HCJ, Douglas AE (2004) Linking the bacterial community in pea aphids with host-plant use and natural enemy resistance. Ecol Entomol 29:60–65. https://doi.org/10.1111/j.1365-2311.2004.00574.x
Oh D-C, Poulsen M, Currie CR, Clardy J (2009) Dentigerumycin: a bacterial mediator of an ant-fungus symbiosis. Nat Chem Biol 5:391–393. https://doi.org/10.1038/nchembio.159
Douglas AE (2015) Multiorganismal insects: diversity and function of resident microorganisms. Annu Rev Entomol 60:17–34. https://doi.org/10.1146/annurev-ento-010814-020822
Flórez LV, Biedermann PHW, Engl T, Kaltenpoth M (2015) Defensive symbioses of animals with prokaryotic and eukaryotic microorganisms. Nat Prod Rep 32:904–936. https://doi.org/10.1039/c5np00010f
Kaltenpoth M, Göttler W, Herzner G, Strohm E (2005) Symbiotic bacteria protect wasp larvae from fungal infestation. Curr Biol 15:475–479. https://doi.org/10.1016/j.cub.2004.12.084
Kaltenpoth M (2009) Actinobacteria as mutualists: general healthcare for insects? Trends Microbiol 17:529–535. https://doi.org/10.1016/j.tim.2009.09.006
Scarborough CL, Ferrari J, Godfray HCJ. Aphid protected from pathogen by endosymbiont. Science (80- ). 2005;310:1781
Mahadav A, Gerling D, Gottlieb Y, Czosnek H, Ghanim M (2008) Parasitization by the wasp Eretmocerus mundus induces transcription of genes related to immune response and symbiotic bacteria proliferation in the whitefly Bemisia tabaci. BMC Genomics 9:342. https://doi.org/10.1186/1471-2164-9-342
Dillon RJ, Dillon VM (2004) The gut bacteria of insects: nonpathogenic interactions. Annu Rev Entomol 49:71–92. https://doi.org/10.1146/annurev.ento.49.061802.123416
Krishnan M, Bharathiraja C, Pandiarajan J, Prasanna VA, Rajendhran J, Gunasekaran P (2014) Insect gut microbiome - an unexploited reserve for biotechnological application. Asian Pac J Trop Biomed 4(Suppl 1):S16–S21. https://doi.org/10.12980/APJTB.4.2014C95
Lewis Z, Lizé A (2015) Insect behaviour and the microbiome. Curr Opin Insect Sci 9:86–90. https://doi.org/10.1016/J.COIS.2015.03.003
Douglas AE (2011) Lessons from studying insect symbioses. Cell Host Microbe 10:359–367. https://doi.org/10.1016/J.CHOM.2011.09.001
Zucchi TD, Prado SS, Cônsoli FL (2012) The gastric caeca of pentatomids as a house for actinomycetes. BMC Microbiol 12:101. https://doi.org/10.1186/1471-2180-12-101
Salem H, Kreutzer E, Sudakaran S, Kaltenpoth M (2013) Actinobacteria as essential symbionts in firebugs and cotton stainers (Hemiptera, Pyrrhocoridae). Environ Microbiol 15:1956–1968. https://doi.org/10.1111/1462-2920.12001
Moran NA, Tran P, Gerardo NM (2005) Symbiosis and insect diversification: an ancient symbiont of sap-feeding insects from the bacterial phylum Bacteroidetes. Appl Environ Microbiol 71:8802–8810. https://doi.org/10.1128/AEM.71.12.8802-8810.2005
Koga R, Bennett GM, Cryan JR, Moran NA (2013) Evolutionary replacement of obligate symbionts in an ancient and diverse insect lineage. Environ Microbiol 15:2073–2081. https://doi.org/10.1111/1462-2920.12121
Koga R, Moran NA (2014) Swapping symbionts in spittlebugs: evolutionary replacement of a reduced genome symbiont. ISME J 8:1237–1246. https://doi.org/10.1038/ismej.2013.235
Scopel W, Cônsoli FL. Culturable symbionts associated with the reproductive and digestive tissues of the neotropical brown stinkbug Euschistus heros. Antonie Van Leeuwenhoek. 2018;:1–12. https://doi.org/10.1007/s10482-018-1130-9
Muyzer G, de Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700 http://www.ncbi.nlm.nih.gov/pubmed/7683183.
Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M et al (2013) Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41:e1–e1. https://doi.org/10.1093/nar/gks808
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. https://doi.org/10.1038/nmeth.f.303
Pylro VS, Roesch LFW, Morais DK, Clark IM, Hirsch PR, Tótola MR (2014) Data analysis for 16S microbial profiling from different benchtop sequencing platforms. J Microbiol Methods 107:30–37. https://doi.org/10.1016/J.MIMET.2014.08.018
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 26:2460–2461. https://doi.org/10.1093/bioinformatics/btq461
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K et al (2006) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. https://doi.org/10.1128/AEM.03006-05
R Core Team. R: Language And Environment For Statistical Computing R Foundation for Statistical Computing. 2017
Ferreira EB, Cavalcanti PP, Nogueira DA (2014) ExpDes: an R package for ANOVA and experimental designs. Appl Math 05:2952–2958. https://doi.org/10.4236/am.2014.519280
McMurdie PJ, Holmes S (2014) Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol 10:e1003531. https://doi.org/10.1371/journal.pcbi.1003531
Dhariwal A, Chong J, Habib S, King IL, Agellon LB, Xia J (2017) MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res 45:W180–W188. https://doi.org/10.1093/nar/gkx295
Kroiss J, Kaltenpoth M, Schneider B, Schwinger M-G, Hertweck C, Maddula RK et al (2010) Symbiotic streptomycetes provide antibiotic combination prophylaxis for wasp offspring. Nat Chem Biol 6:261–263. https://doi.org/10.1038/nchembio.331
Marshall AT (1965) Batelli glands of cercopoid nymphs (Homoptera). Nature. 205:925
Cecil R (1930) The alimentary canal of Philaenus leucophthalmus L. Ohio J Sci 30:120–130
Wilson HA, Dorsey CK (1957) Studies on the composition and microbiology of insect spittle. Ann Entomol Soc Am 50:399–406. https://doi.org/10.1093/aesa/50.4.399
Ben BC, Cordon-Rosales C, Durvasula RV (2002) Bacterial symbionts of the triatominae and their potential use in control of Chagas disease transmission. Annu Rev Entomol 47:123–141. https://doi.org/10.1146/annurev.ento.47.091201.145144
Hosokawa T, Kikuchi Y, Nikoh N, Shimada M, Fukatsu T (2006) Strict host-symbiont cospeciation and reductive genome evolution in insect gut bacteria. PLoS Biol 4:e337. https://doi.org/10.1371/journal.pbio.0040337
Prado SS, Rubinoff D, Almeida RPP (2006) Vertical transmission of a pentatomid caeca-associated symbiont. Ann Entomol Soc Am 99:577–585. https://doi.org/10.1603/0013-8746(2006)99[577:vtoapc]2.0.co;2
Park D-S, Oh H-W, Jeong W-J, Kim H, Park H-Y, Bae KS (2007) A culture-based study of the bacterial communities within the guts of nine longicorn beetle species and their exo-enzyme producing properties for degrading xylan and pectin. J Microbiol 45:394–401 http://www.ncbi.nlm.nih.gov/pubmed/17978798.
Lefebvre T, Miambi E, Pando A, Diouf M, Rouland-Lefèvre C (2009) Gut-specific actinobacterial community structure and diversity associated with the wood-feeding termite species, Nasutitermes corniger (Motschulsky) described by nested PCR-DGGE analysis. Insect Soc 56:269–276. https://doi.org/10.1007/s00040-009-0020-6
Hedges LM, Brownlie JC, O’Neill SL, Johnson KN (2008) Wolbachia and virus protection in insects. Science. 322:702. https://doi.org/10.1126/science.1162418
Funding
This work was supported in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Brazil (CAPES), Finance Code 001, and the National Institute of Science and Technology – Semiochemicals in Agriculture (Fundação de Amparo à Pesquisa do Estado de São Paulo and Conselho Nacional de Desenvolvimento Científico e Tecnológico [grants #2014/50871-0 and #465511/2014-7, respectively]).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Responsible Editor: Lucy Seldin.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 19 kb)
Rights and permissions
About this article
Cite this article
Tonelli, M., Cotta, S.R., Rigotto, A. et al. The composition of the bacterial community in the foam produced by Mahanarva fimbriolata is distinct from those at gut and soil. Braz J Microbiol 51, 1151–1157 (2020). https://doi.org/10.1007/s42770-019-00211-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-019-00211-1