Abstract
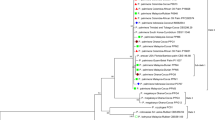
The genus Xanthomonas comprises Gram-negative bacteria, many of which are phytopathogens. Xanthomonas fuscans subsp. fuscans is one of the most devastating pathogens affecting the bean plant, resulting in the common bacterial blight of bean (CBB). The disease is mainly foliar and affects a wide variety of bean species, thus acting as the yield-limiting factor for the bean crop. Here, we report the whole-genome sequencing of a new strain of X. fuscans subsp. fuscans, named Xff49, isolated from the infected and symptomatic beans from Capão do Leão, Southern Brazil. The genetic analysis demonstrated the presence of single-nucleotide variants (SNVs) in this strain, potentially affecting the mobilome, cell mobility, and inorganic ion metabolism. In addition, the analysis resulted in the identification of a new plasmid similar to the pAX22 derived from Achromobacter denitrificans, which was named plX, along with plA and plC, previously reported in other strains of X. fuscans subsp. fuscans. Xff49 represents the first Brazilian genome of X. fuscans subsp. fuscans and might provide useful information applicable to the studies of phylogenetics, evolution, and pathogenomics, thereby allowing a better understanding of the genomic features present in the Brazilian strains.





Similar content being viewed by others
References
Ryan RP, Vorhölter F-J, Potnis N, Jones JB, van Sluys MA, Bogdanove AJ, Dow JM (2011) Pathogenomics of Xanthomonas: understanding bacterium–plant interactions. Nat Rev Microbiol 9(5):344–355. https://doi.org/10.1038/nrmicro2558
Akhavan A, Bahar M, Askarian H, Lak MR, Nazemi A, Zamani Z (2013) Bean common bacterial blight: pathogen epiphytic life and effect of irrigation practices. Springerplus. 2(1):41. https://doi.org/10.1186/2193-1801-2-41.
Morris CE, Monier J-M (2003) The ecological significance of biofilm formation by plant-associated bacteria. Annu Rev Phytopathol 41(1):429–453. https://doi.org/10.1146/annurev.phyto.41.022103.134521
Graham PH, Vance CP (2003) Legumes: Importance and constraints to greater use. Plant Physiol 131(3):872–877. https://doi.org/10.1104/pp.017004.
Shi C, Navabi A, Yu K (2011) Association mapping of common bacterial blight resistance QTL in Ontario bean breeding populations. BMC Plant Biol 11(1):52. https://doi.org/10.1186/1471-2229-11-52
Durham KM, Xie W, Yu K, Pauls KP, Lee E, Navabi A (2013) Interaction of common bacterial blight quantitative trait loci in a resistant inter-cross population of common bean. Link W, ed. Plant Breed 132(6):658–666. https://doi.org/10.1111/pbr.12103
Zhu J, Wu J, Wang L, Blair MW, Zhu Z, Wang S (2016) QTL and candidate genes associated with common bacterial blight resistance in the common bean cultivar Longyundou 5 from China. Crop J 4(5):344–352. https://doi.org/10.1016/j.cj.2016.06.009
Jacques M-A, Josi K, Darrasse A, Samson R (2005) Xanthomonas axonopodis pv. phaseoli var. fuscans is aggregated in stable biofilm population sizes in the phyllosphere of field-grown beans. Appl Environ Microbiol 71(4):2008–2015. https://doi.org/10.1128/AEM.71.4.2008-2015.2005
Ruh M, Briand M, Bonneau S, Jacques M-A, Chen NWG (2017) Xanthomonas adaptation to common bean is associated with horizontal transfers of genes encoding TAL effectors. BMC Genomics 18(1):670. https://doi.org/10.1186/s12864-017-4087-6.
Darrasse A, Carrère S, Barbe V, Boureau T, Arrieta-Ortiz ML, Bonneau S, Briand M, Brin C, Cociancich S, Durand K, Fouteau S, Gagnevin L, Guérin F, Guy E, Indiana A, Koebnik R, Lauber E, Munoz A, Noël LD, Pieretti I, Poussier S, Pruvost O, Robène-Soustrade I, Rott P, Royer M, Serres-Giardi L, Szurek B, van Sluys MA, Verdier V, Vernière C, Arlat M, Manceau C, Jacques MA (2013) Genome sequence of Xanthomonas fuscans subsp. fuscans strain 4834-R reveals that flagellar motility is not a general feature of xanthomonads. BMC Genomics 14:761. https://doi.org/10.1186/1471-2164-14-761
Indiana A, Briand M, Arlat M, Gagnevin L, Koebnik R, Noel LD, Portier P, Darrasse A, Jacques MA (2014) Draft genome sequence of the flagellated Xanthomonas fuscans subsp. fuscans strain CFBP 4884. Genome Announc 2(5). https://doi.org/10.1128/genomeA.00966-14
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Tritt A, Eisen JA, Facciotti MT, Darling AE (2012) An integrated pipeline for de novo assembly of microbial genomes. PLoS One 7(9):e42304. https://doi.org/10.1371/journal.pone.0042304
Zerbino DR, Birney E (2008) Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18(5):821–829. https://doi.org/10.1101/gr.074492.107
Simpson JT, Durbin R (2012) Efficient de novo assembly of large genomes using compressed data structures. Genome Res 22(3):549–556. https://doi.org/10.1101/gr.126953.111
Boisvert S, Laviolette F, Corbeil J (2010) Ray: simultaneous assembly of reads from a mix of high-throughput sequencing technologies. J Comput Biol 17(11):1519–1533. https://doi.org/10.1089/cmb.2009.0238
Lin SH, Liao YC (2013) CISA: contig integrator for sequence assembly of bacterial genomes. PLoS One 8(3):1–7. https://doi.org/10.1371/journal.pone.0060843
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: architecture and applications. BMC Bioinformatics 10(1):421. https://doi.org/10.1186/1471-2105-10-421
Bosi E, Donati B, Galardini M, Brunetti S, Sagot MF, Lió P, Crescenzi P, Fani R, Fondi M (2015) MeDuSa: a multi-draft based scaffolder. Bioinformatics. 31(15):2443–2451. https://doi.org/10.1093/bioinformatics/btv171
Lu CL, Chen KT, Huang SY, Chiu HT (2014) Car: Contig assembly of prokaryotic draft genomes using rearrangements. BMC Bioinformatics 15(1):1–10. https://doi.org/10.1186/s12859-014-0381-3
Tsai IJ, Otto TD, Berriman M (2010) Improving draft assemblies by iterative mapping and assembly of short reads to eliminate gaps. Genome Biol 11(4):R41. https://doi.org/10.1186/gb-2010-11-4-r41
Gurevich A, Saveliev V, Vyahhi N, Tesler G (2013) QUAST: quality assessment tool for genome assemblies. Bioinformatics. 29(8):1072–1075. https://doi.org/10.1093/bioinformatics/btt086
Kremer FS, Eslabão MR, Dellagostin OA, Pinto L d S (2016) Genix: a new online automated pipeline for bacterial genome annotation. FEMS Microbiol Lett 11(16)
Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B (2000) Artemis: sequence visualization and annotation. Bioinformatics. 16(10):944–945. https://doi.org/10.1093/bioinformatics/16.10.944
Wang Y, Coleman-Derr D, Chen G, Gu YQ (2015) OrthoVenn: a web server for genome wide comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res 43(W1):W78–W84. https://doi.org/10.1093/nar/gkv487
Blin K, Wolf T, Chevrette MG, Lu X, Schwalen CJ, Kautsar SA, Suarez Duran HG, de los Santos ELC, Kim HU, Nave M, Dickschat JS, Mitchell DA, Shelest E, Breitling R, Takano E, Lee SY, Weber T, Medema MH (2017) antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res 45(W1):W36–W41. https://doi.org/10.1093/nar/gkx319
Angiuoli SV, Salzberg SL (2011) Mugsy: fast multiple alignment of closely related whole genomes. Bioinformatics. 27(3):334–342. https://doi.org/10.1093/bioinformatics/btq665
Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973. https://doi.org/10.1093/bioinformatics/btp348
Cock PJA, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, de Hoon MJL (2009) Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 25(11):1422–1423. https://doi.org/10.1093/bioinformatics/btp163
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup (2009) The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079. https://doi.org/10.1093/bioinformatics/btp352
Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w 1118; iso-2; iso-3. Fly (Austin) 6(2):80–92. https://doi.org/10.4161/fly.19695.
Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Bryant SH (2014) CDD: NCBI’s conserved domain database. Nucleic Acids Res 43(Database issue):D222–D226. https://doi.org/10.1093/nar/gku1221
Tatusov RL (2000) The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28(1):33–36. https://doi.org/10.1093/nar/28.1.33
Moreira LM, Almeida NF, Potnis N, Digiampietri LA, Adi SS, Bortolossi JC, da Silva AC, da Silva AM, de Moraes FE, de Oliveira JC, de Souza RF, Fancincani AP, Ferraz AL, Ferro MI, Furlan LR, Gimenez DF, Jones JB, Kitajima EW, Laia ML, Leite RP, Nishiyama MY, Rodrigues Neto J, Nociti LA, Norman DJ, Ostroski EH, Pereira HA, Staskawicz BJ, Tezza RI, Ferro JA, Vinatzer BA, Setubal JC (2010) Novel insights into the genomic basis of citrus canker based on the genome sequences of two strains of Xanthomonas fuscans subsp. aurantifolii. BMC Genomics 11(1):238. https://doi.org/10.1186/1471-2164-11-238
Abendroth U, Schmidtke C, Bonas U (2014) Small non-coding RNAs in plant-pathogenic Xanthomonas spp. RNA Biol 11(5):457–463. https://doi.org/10.4161/rna.28240
Schmidtke C, Abendroth U, Brock J, Serrania J, Becker A, Bonas U, Small RNA (2013) sX13: a multifaceted regulator of virulence in the plant pathogen Xanthomonas. Waldor MK, ed. PLoS Pathog 9(9):e1003626. https://doi.org/10.1371/journal.ppat.1003626.
Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR (2003) Rfam: an RNA family database. Nucleic Acids Res 31(1):439–441 http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=165453&tool=pmcentrez&rendertype=abstract. Accessed 26 Mar 2015
Nawrocki EP, Kolbe DL, Eddy SR (2009) Infernal 1.0: inference of RNA alignments. Bioinformatics. 25(10):1335–1337. https://doi.org/10.1093/bioinformatics/btp157
Zdobnov EM, Apweiler R (2001) InterProScan--an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 17(9):847–848 http://www.ncbi.nlm.nih.gov/pubmed/11590104. Accessed 12 May 2014
Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer ELL, Tate J, Punta M (2014) Pfam: the protein families database. Nucleic Acids Res 42(Database issue):D222–D230. https://doi.org/10.1093/nar/gkt1223
Letunic I, Doerks T, Bork P (2012) SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res 40(Database issue):D302–D305. https://doi.org/10.1093/nar/gkr931
Buttimer C, McAuliffe O, Ross RP, Hill C, O’Mahony J, Coffey A (2017) Bacteriophages and bacterial plant diseases. Front Microbiol 8:34. https://doi.org/10.3389/fmicb.2017.00034
Hu Z, Dong H, Ma J-C, Yu Y, Li KH, Guo QQ, Zhang C, Zhang WB, Cao X, Cronan JE, Wang H (2018) Novel Xanthomonas campestris long-chain-specific 3-Oxoacyl-acyl carrier protein reductase involved in diffusible signal factor synthesis. Lindow SE, ed. MBio 9(3). https://doi.org/10.1128/mBio.00596-18.
Nikaido H, Takatsuka Y (2009) Mechanisms of RND multidrug efflux pumps. Biochim Biophys Acta, Proteins Proteomics 1794(5):769–781. https://doi.org/10.1016/j.bbapap.2008.10.004
Brakhage AA (1998) Molecular regulation of beta-lactam biosynthesis in filamentous fungi. Microbiol Mol Biol Rev 62(3):547–585 http://www.ncbi.nlm.nih.gov/pubmed/9729600. Accessed 7 Oct 2018
Wright GD (2010) Antibiotic resistance in the environment: a link to the clinic? Curr Opin Microbiol 13(5):589–594. https://doi.org/10.1016/j.mib.2010.08.005
Pandey SS, Patnana PK, Rai R, Chatterjee S (2017) Xanthoferrin, the α-hydroxycarboxylate-type siderophore of Xanthomonas campestris pv. campestris , is required for optimum virulence and growth inside cabbage. Mol Plant Pathol 18(7):949–962. https://doi.org/10.1111/mpp.12451
Bianco MI, Toum L, Yaryura PM, Mielnichuk N, Gudesblat GE, Roeschlin R, Marano MR, Ielpi L, Vojnov AA (2016) Xanthan pyruvilation is essential for the virulence of Xanthomonas campestris pv. campestris. Mol Plant-Microbe Interact 29(9):688–699. https://doi.org/10.1094/MPMI-06-16-0106-R
Poplawsky AR, Urban SC, Chun W (2000) Biological role of Xanthomonadin pigments in Xanthomonas campestris pv. campestris. Appl Environ Microbiol 66(12):5123. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC92432/. Accessed 11 June 2018–5127
Huang C-L, Pu P-H, Huang H-J, Sung HM, Liaw HJ, Chen YM, Chen CM, Huang MB, Osada N, Gojobori T, Pai TW, Chen YT, Hwang CC, Chiang TY (2015) Ecological genomics in Xanthomonas: the nature of genetic adaptation with homologous recombination and host shifts. BMC Genomics 16(1):188. https://doi.org/10.1186/s12864-015-1369-8.
Funding
This work was financially supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Brazil).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Rodrigo Galhardo.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kremer, F.S., de Souza Junior, I.T., Guimarães, A.M. et al. Genome sequence of Xanthomonas fuscans subsp. fuscans strain Xff49: a new isolate obtained from common beans in Southern Brazil. Braz J Microbiol 50, 357–367 (2019). https://doi.org/10.1007/s42770-019-00050-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-019-00050-0