Abstract
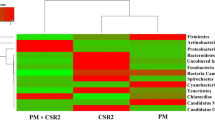
Information on the bacterial symbiosis/interactions is lacking in tasar silkworm Antheraea mylitta Drury. The detailed midgut bacterial diversity and their functions were elucidated for the first time in tasar silkworm. Taxonomic annotation of fifth instar larval midgut bacteria of three major ecoraces namely Daba, Modal and Raily was carried out using Illumina Next-Generation Sequencing. The dominant phyla and genus are Proteobacteria and Pseudomonas respectively, irrespective of the ecorace. The bacterial diversity & functions varied among ecoraces due to diet & geography. Predictive functional analysis through PICRUSt, revealed many important functional roles of gut bacteria in digestion, detoxification, amino-acid synthesis, etc. Metabolic pathways related to metabolism of amino acids, carbohydrates, lipids, plant secondary metabolites and membrane transport are the most significant gut bacterial functions. Loss of native gut bacteria following the antibiotics treatment reduced the silkworm fitness. We found significant symbiotic role between silkworm and midgut bacteria mediated through various physiological functions. The information obtained from this study helps to enrich the knowledge and could contribute to engineer the functions for the enhancement of silk yield and quality through probiotics.




Similar content being viewed by others
References
Adams AS, Aylward FO, Adams SM, Erbilgin N, Aukema BH, Currie CR, Suen G, Raffa KF (2013) Mountain pine beetles colonizing historical and naïve host trees are associated with a bacterial community highly enriched in genes contributing to terpene metabolism. Appl Environ Microbiol 79:3468–75
Almeida LGd, Moraes LABd, Trigo JR, Omoto C, Consoli FL (2017) The gut microbiota of insecticide resistant insect houses insecticide degrading bacteria: A potential source for biotechnological exploitation. PLoS ONE 12(3):e0174754
Bansal R, Hulbert SH, Reese JC, Whitworth RJ, Stuart JJ, Chen MS (2014) Pyrosequencing reveals the predominance of Pseudomonadaceae in gut microbiome of a gall midge. Pathogens 3(2):459–72
Bapatla KG, Singh A, Yeddula S, Patil RH (2018) Annotation of gut bacterial taxonomic and functional diversity in Spodoptera litura and Spilosoma obliqua. J Basic Microbiol 58(3):217–26
Bennett GM, McCutcheon JP, MacDonald BR, Romanovicz D, Moran NA (2014) Differential genome evolution between companion symbionts in an insect-bacterial symbiosis. mBio 5:e01697
Berasategui A, Salem H, Paetz C, Santoro M, Gershenzon J, Kaltenpoth M, Schmidt A (2017) Gut microbiota of the pine weevil degrades conifer diterpenes and increases insect fitness. Mol Ecol 00:1–12
Briones-Roblero CI, HernaÂndez-GarcõÂa JA, Gonzalez-Escobedo R, Soto-Robles LV, Rivera- Orduña FN, ZuÂñiga G (2017) Structure and dynamics of the gut bacterial microbiota of the bark beetle, Dendroctonus rhizophagus (Curculionidae: Scolytinae) across their life stages. PLoS ONE 12(4):e0175470
Broderick NA, Raffa KF, Goodman RM, Handelsman J (2004) Census of the bacterial community of the gypsy moth larval midgut by using culturing and culture-independent methods. Appl Environ Microbiol 70(1):293–300
Brown SD, Lamed R, Morag E, Borovok I, Shoham Y, Klingeman DM, Johnson CM, Yang Z, Land ML, Utturkar SM, Keller M, Bayer EA (2012) Draft genome sequences for Clostridium thermocellum wild-type strain YS and derived cellulose adhesion-defective mutant strain AD2. J. Bacteriol 194:3290–1
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allowsanalysis of high-throughput community sequencing data. Nat Methods 7(5):335–6
Ceja-Navarro JA, Vega FE, Karaoz U, Hao Z, Jenkins S, Lim HC, Kosina P, Infante F, Northen TR, Brodie EL (2015) Gut microbiota mediatecaffeine detoxification in the primary insect pest of coffee. Nat Commun 6:7618
Chen B, Zhanga N, Xiea S, Zhanga X, Hea J, Muhammada A, Sun Lua X, Shaoa Y (2020) Gut bacteria of the silkworm Bombyx mori facilitate host resistance against the toxic effects of organophosphate insecticides. Environ Int 143:105886
Chen B, Yu T, Xie S, Du K, Liang X, Lan Y, Sun C, Lu X, Shao Y (2018) Comparative shotgun metagenomic data of the silkworm Bombyx mori gut microbiome. Sci Data 5:180285
CTR&TI (2020-2021) Annual report. Central Tasar Research and Training Institute, Ranchi, Jharkhand, Central Silk Board, India
Delalibera JR, Handelsman J, Raffa KF (2005) Contrasts in cellulolytic activities of gutmicroorganisms between the wood borer, Saperda vestita (Coleoptera:Cerambycidae), and the bark beetles, Ips pini and Dendroctonus frontalis (Coleoptera: Curculionidae). Environ Entomol 34:541–7
DeSantis TZ Jr, Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM, Phan R, Andersen GL (2006a) NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res 34:394–9
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL (2006b) Greengenes, a Chimera-Checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069-72
Deutscher AT, Burke CM, Darling AE, Riegler M, Reynolds OL, Chapman TA (2018) Near full-length 16S rRNA gene next-generation sequencing revealed Asaiaas a common midgut bacterium of wild and domesticated Queensland fruitfly larvae. Microbiome 6:85
Dillon RJ, Dillon VM (2004) The gut bacteria of insects: nonpathogenic interactions.Annu Rev Entomol 49:71–92
Dong HL, Zhang SX, Chen ZH, Tao H, Li X,Qiu JF, Cui WZ, Sima YH, Cui WZ, Xu SQ (2018) Differences in gut microbiota between silkworms Bombyx mori reared on fresh mulberry (Morusalba var. multicaulis) leaves or an artificial diet. RSC Adv 8:26188
Douglas AE (2015) The multi-organismal insect: diversity and function of resident microorganisms. Annu Rev Entomol 60:17–34
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–1
Engel P, Moran NA (2013) The gut microbiota of insects - diversity in structure and function. FEMS Microbiol Rev 37:699–735
Gandotra S, Kumar A, Naga K, Bhuyan PM, Gogoi DK, Sharma K, Subramanian S (2018) Bacterial community structure and diversity in the gut of the muga silkworm, Antheraea assamensis (Lepidoptera: Saturniidae), from India. Insect Mol Biol 27(5):603–19
Gavriel S, Jurkevitch E, Gazit Y, Yuval B (2010) Bacterially enriched diet improves sexual performance of sterile male Mediterranean fruit flies. J Appl Entomol. https://doi.org/10.1111/j.1439-0418.2010.01605.x
Hammer TJ, Janzen DH, Hallwachs W, Jaffe SP, Fierer N (2017) Caterpillars lack a resident gut microbiome. Proc Natl Acad Sci 114(36):9641–46
Indiragandhi P, Anandham R, Madhaiyan M, Sa TM (2008) Characterization of plant growth-promoting traits of bacteria isolated from larval guts of diamondback moth Plutella xylostella (Lepidoptera: Plutellidae). Curr Microbiol 56:327–33
Jolly MS, Sen SK, Sonwalkaar TN, Prasad GK (1979) Non-Mulberry Silks. FAO Agricultural Service Bulletin no. 4. FAO, Rome
Jovel J, Patterson J, Wang W, Hotte N, O’Keefe S, Mitchel T, Perry T, Kao D, Mason AL, Madsen KL, Wong GK-S (2016) Characterization of the gut microbiome using 16S or shotgun metagenomics. Front Microbiol 7:459
Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31:814–21
Liang X, Fu Y, Tong L, Liu H (2014) Microbial shifts of the silkworm larval gut in response to lettuce leaf feeding. Appl Microbiol Biotechnol 98:3769–76
Lu¨ J, Guo W, Chen S, Guo M, Qiu B, Yang C, Lian T, Pan H (2019) Host plants influence the composition of the gut bacteria in Henosepilachna vigintioctopunctata. PLoS ONE 14(10): e0224213
Manjappa Seema Bara, Prabhu Immanual Gilwax, Baig Mohammed Muzeruddin, Sahay Alok (2020) Influence of Shorea robusta leaf extract treatment on Terminalia arjuna plants over tasar silkworm growth and economic traits. J Entomol Zool Stud 8(3):1095–101
Martínez-Solís M, Collado MC, Herrero S (2020) Influence of diet, sex, and viral infections on the gut microbiota composition of Spodoptera exigua caterpillars. Front. Microbiol 11:753
Masona J, Raya S, Shikanoa I, Peiffera M, Jonesa AG, Lutheb DS, Hoovera K, Feltona GW (2019) Plant defences interact with insect enteric bacteria by initiating a leaky gut syndrome Charles. PNAS 116(32):15991–6
Mazid M, Khan T, Mohammad F (2011) Role of secondary metabolites indefense mechanisms of plants. Biol Med 3:232–49
MsangoSoko K, Gandotra S, Chandel R, Sharma K, Ramakrishinan B, Subramanian S (2020) Composition and diversity of gut bacteria associated with the eri silk moth, Samia ricini, (Lepidoptera: Saturniidae) as revealed by culture-dependent and metagenomics analysis. J Microbiol Biotechnol 30:10
Paniagua Voirol LR, Frago E, Kaltenpoth M, Hilker M, Fatouros NE (2018) Bacterial Symbionts in Lepidoptera: Their Diversity, Transmission, and Impact on the Host. Front Microbiol 9:556
Pinto-Toma´ S, Ana Sittenfeld, Lorena Uribe-Lori´O, Felipe Chavarri´A, Marielos Mora, Daniel H. Janzen, Robert M. Goodman, Holly MS (2011) Comparison of midgut bacterial diversity in tropical caterpillars (Lepidoptera: Saturniidae) fed on different diets. Environ Entomol 40(5):1111-22
Priya NG, Ojha A, Kajla MK, Raj A, Rajagopal R (2012) Host plant induced variation in gut bacteria of Helicoverpa armigera. PLoS One 7:e30768
Qualley AV, Widhalm JR, Adebesin F, Kish CM, Dudareva N (2012) Completion of the core b-oxidative pathway of benzoic acidbiosynthesis in plants. Proc Natl Acad Sci 109:16383–8
Rani A, Sharma A, Rajagopal R, Adak T, Bhatnagar RK (2009) Bacterial diversity analysis of larvae and adult midgut microflora using culture-dependent andculture-independent methods in lab-reared and field-collected Anopheles stephensi - an Asian malarial vector. BMC Microbiol 9:96
Rajan Resma, Chanda Shampa Deb, Rani Alekhya, Gattu Renuka, Vodithala Shamitha, Mamillapalli Anitha (2020) Bacterial gut symbionts of Antheraea mylitta (Lepidoptera: Saturniidae). J Entomolo Sci 55(1):137–40
Schrempf H (2001) Recognition and degradation of chitin by Streptomycetes. Anton Leeuw Int J G 79:285–9
Senderovich Y, Halpern M (2013) The protective role of endogenous bacterial communities in chironomid egg masses and larvae. ISME J 7:2147–58
Sharon G, Segal D, Ringo JM, Hefetz A, Rosenberg IZ, Rosenberg E (2010) Commensal bacteria play a role in mating preference of Drosophila melanogaster. PNAS 107(46):20051–56
Staudacher H, Kaltenpoth M, Breeuwer JAJ, Menken SBJ, Heckel DG, Groot AT (2016) Variability of bacterial communities in the moth Heliothis virescens indicates transient association with the host. PLoS ONE 11(5):e0154514
Vernier CL, Chin IM, Adu-Oppong B, Krupp JJ, Levine J, Dantas G, Ben-Shahar Y (2020) The gut microbiome defines social group membership in honey bee colonies. Sci Adv 6: eabd3431
Visotto LE, Oliviera MGA, Guedes RNC, Ribon AOB, Good-God PIV (2009) Contribution of gut bacteria to digestion and development of the velvetbean caterpillar, Anticarsia gemmatalis. J Insect Physiol 55:185–91
Werren JH (2012) Symbionts provide pesticide detoxification. Proc Natl Acad Sci 109:8364–5
Xia H, Chunyan W, Hui C, Junning M (2013a) Difference in the structure of the gut bacteria communities in developmental stages of the Chinese white pine beetle (Dendroctonus armandi). Int J Mol Sci 14:21006–20
Xia X, Gurr GM, Vasseur L, Zheng D, Zhong H, Qin B, Lin J, Wang Y, Song F, Li Y, Lin H, You M (2017) Metagenomic sequencing ofdiamondback moth gut microbiomeunveils key holobiont adaptations forherbivory. Front Microbiol 8:663
Xia X, Lan B, Tao X, Lin J, You M (2020) Characterization of Spodoptera litura gut bacteria and their role in feeding and growth of the host. Front Microbiol 11:1492
Xia X, Sun B, Gurr GM, Vasseur L, Xue M, You M (2018) Gut Microbiota Mediate Insecticide Resistance in the Diamondback Moth, Plutella xylostella (L.). Front Microbiol 9:25
Xia, X, Zheng D, Zhong H, Qin B, Gurr GM, Vasseur L, Lin H, Bai J, He W, You M (2013b) DNA sequencing reveals the midgut microbiota of diamondback moth, Plutella xylostella (L.) and a possible relationship with insecticide resistance. PLoS ONE 8(7):e68852
Xu B, Xu W, Yang F, Li J, Yang Y, Tang X, Mu Y, Zhou J, Huang Z (2013) Metagenomic analysis of the Pygmy loris fecal microbiome reveals unique functional capacity related to metabolism of aromatic compounds. PLoS ONE 8:e56565
Yun JH, Roh SW, Whon TW, Jung MJ, Kim MS, Park DS, Yoon C, Nam YD, Kim YJ, Choi JH, Kim JY, Shin NR, Kim SH, Lee WJ, Bae JW (2014) Insect gut bacterial diversity determined by environmental habitat, diet, developmental stage, and phylogeny of host. Appl Environ Microbiol 80:5254–64
Acknowledgments
Present study was carried out with the funds of Central Silk Board (Code: PIP 4716, CTRTI/PMEC/TECH-60/2016-17/8846). We thank Dr. P.K. Kar, Scientist-D, Baripada, CSB for helping in the procurement of Modal ecorace larvae. We thank Dr. Jaipal Choudhary, Scientist, ICAR-RCER, Ranchi & Dr. Rahul Benerjee, Scientist, ICAR-IASRI, New Delhi for their valuable help in bioinformatics.
Author information
Authors and Affiliations
Contributions
MMB and Alok conceived the project. MMB designed and performed the experiments, analysed the metagenomic data and wrote the manuscript. Prabhu & Manjappa helped in the investigation and methodology. Singh & Sathyanarayana provided the necessary inputs during the experiments. All the authors gave final approval for the manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors have declared no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
42690_2023_1006_MOESM1_ESM.docx
Supplementary file1 Life cycle of Tasar Silkworm Antheraea mylitta Drury. a: mating, b: Hatching, c-f: II, III, IV and V instar larvae respectively, g: Cocoon with peduncle, h: Adult moth (DOCX 16 KB)
42690_2023_1006_MOESM2_ESM.jpg
Supplementary file2 The phylogenetic tree of A.mylitta larval midgut bacteria. Phylogenetic tree with seed value of 100 and bootstrapping value of 1000. a. Modal, b. Raily, c. Daba (JPG 1063 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Baig, M.M., Singh, G., Prabhu, D.I.G. et al. Characterization of tasar silkworm Antheraea mylitta drury (Saturniidae: Lepidoptera) midgut bacterial symbionts through metagenomic analysis. Int J Trop Insect Sci 43, 999–1011 (2023). https://doi.org/10.1007/s42690-023-01006-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42690-023-01006-6